International Journal of Agricultural Science and Food Technology
Genetic variability, heritability, and genetic advance for quantitative traits of sorghum [Sorghum Bicolor (L.) Moench] genotypes at Fedis, Eastern Ethiopia
Mohammed Jafar1*, Bulti Tesso2 and Girma Mengistu3
2School of Plant Sciences, College of Agriculture and Environmental Sciences, Haramaya University, Haramaya, Ethiopia
3Oromia Agricultural Research Institute, Finfinne, Ethiopia
Cite this as
Jafar M, Tesso B, Mengistu G (2023) Genetic variability, heritability, and genetic advance for quantitative traits of sorghum [Sorghum Bicolor (L.) Moench] genotypes at Fedis, Eastern Ethiopia. Int J Agric Sc Food Technol 9(3): 064-075. DOI: 10.17352/2455-815X.000195Copyright License
© 2023 Jafar M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Sorghum is the second most important food crop after teff in Ethiopia. The objective of the study was to estimate the genetic variations, heritability, and expected genetic advances in the selected sorghum genotypes. Sixty-four sorghum genotypes were evaluated for 17 quantitative traits in 8x8 simple lattice designs at the Boko research site. The analysis of variance revealed highly significant differences among genotypes for all traits. The Phenotypic Coefficient Of Variation (PCV) ranged from 4.74% for days to flowering to 24.74% for panicle width, while Genotypic Coefficients Of Variation (GCV) ranged from 3.58% for leaf length to 20.33 % for panicle width. The highest PCV and GCV values were recorded for panicle width, head weight, and harvest index. Moderate PCV and GCV were recorded for grain filling period, grain filling rate, plant height, panicle length, and grain yield; indicating the effectiveness of selection based on the phenotypic performance of the genotypes. Broad sense heritability (H2) ranged from 24.74 % for biomass yield to 96.6% for head weight, whereas GAM ranged from 4.8% for biomass yield to 41.95% for panicle width. High H2 coupled with high GAM was observed for grain filling rate, panicle length, panicle width, head weight, grain yield, and harvest index; indicating that these characters are controlled by additive gene action and phenotypic selection for these characters will be effective. However, the information generated in the current study it can be useful for breeders who want to improve yield and yield contributing traits of sorghum.
Introduction
Sorghum [(Sorghum bicolor (L.) Moench)] is the fifth most important cereal crop in the world after wheat, rice, maize, and barley [1]. It is a widely cultivated cereal crop in the semi-arid tropical regions of Asia, Africa, and Central America [2]. Sorghum is known as a “camel of crops” due to its high tolerance to water and temperature stress [3]. Sorghum is a diploid (2n=2x=20) tropical origin of C4 crop with high photosynthesis efficiency and a monocotyledon plant belonging to the Poaceae family [4]. Ethiopia is the center of origin and diversity for sorghum which indicates the availability of enormous genetic variability in both cultivated and wild sorghum.
The global sorghum production is estimated to be 57.89 million tons from 40 million hectares of land. In Africa, sorghum production is 29.14 million tons from 26.03 hectares of land. Ethiopia is the third largest sorghum producer in Africa next to Nigeria and Sudan [5]. Sorghum ranks third in area coverage, after maize and teff and it accounts for 15.71% of the total annual cereal (88.52%) grain production. The area covered with sorghum is 1.8 million hectares and total production is 4.52 million tons and the national average productivity of sorghum in Ethiopia is 2.69 tons ha-1 [6]. But the potential yield of the crop can be as high as 6 tons ha-1 [5]. Various biotic factors (parasitic weed Striga, diseases, and insect pests) and abiotic factors (drought and low soil fertility) contribute to the low productivity of sorghum in Ethiopia [7]. In the lowland areas of Ethiopia including East Hararghe, the growing season is short; rainfall is also erratic and unreliable. Due to the limited number of early-maturing varieties which have good biomass yield in such areas, the late-maturing sorghum cultivar grown by farmers is frequently exposed to moisture stress at phases of growth that result in either low yield or total crop failure [8].
Due to these problems, in the study area, the current sorghum production per unit area is not sufficient to meet the demand for human consumption, animal feed, fuel, and building material requirements of a rapidly growing population. The development of sorghum varieties for high yield with desirable traits helps in improving food insecurity problems in the area. Genetic improvement in sorghum yield depends on the magnitude of genetic variability, heritability, and genetic advance in the population. In planning a sorghum improvement program, knowledge of the variability of traits could be a key success. Genetic parameters like the genotypic coefficient of variation, phenotypic coefficient of variation, heritability, and genetic advance are useful biometric tools for measuring genetic variability [9]. Success in crop breeding depends on the isolation of genetically superior genotypes based on the amount of variability present in the material. Then, information on genetic variability existing in a set of populations of sorghum is essential. The progress of selection is more important in any crop improvement and this progress depends on the existence of genetic variability for yield and yield contributing characters and their heritability [10]. Heritability in combination with genetic advance has a greater role to play in determining the effectiveness of selection of a character.
Previous studies have indicated the presence of a high level of phenotypic variation in sorghum for quantitative traits among the Ethiopian sorghum collections [11-14]. However, there is limited information on genetic variability in the Ethiopian lowland sorghum lines developed through crossing which is a major concern for sorghum improvement programs. Thus, there is a need for the assessment of genetic variability in advanced sorghum genotypes to increase the efficiency of the breeding program for the target area. Therefore, this study was conducted to estimate the genetic variations, heritability, and expected genetic advances in the selected sorghum genotypes.
Materials and methods
Experimental site
The study was conducted at the Boko research site of Fedis Agricultural Research Center, East Hararghe Zone in the 2021 cropping season. The area is situated at a distance of about 24 km away from Harar town in the southern direction. Fedis is located at the latitude of 09o 07‘North and longitude of 042o 04‘East, and altitude of 1702 meters above sea level, with a prevalence of lowlands. The soil of the experimental site is black with sand clay loam surface soil texture that contains 8.20% organic matter, 0.13% total nitrogen, available phosphorus of 4.99 ppm, soil exchangeable potassium of 1.68 cmol (+) / kg, and a pH value of 8.26. The mean rainfall is about 801.3 mm for the last seven years (2015 to 2021). The mean maximum and minimum annual temperatures are 27.7 and 11.3 °C, respectively, for the last seven years (2015 to 2021) (FARC, 2021).
In this study, 60 sorghum genotypes and two varieties (Argiti and Melkam) were obtained from Melkassa Agricultural Research Center and two varieties (Fedis 01 and Erer) were obtained from Fedis Agricultural Research Center (Table 1). Four released varieties were used as standard checks. The description of the materials is presented in Table 1.
Experimental design and management
The field experiment was laid out in an 8x8 simple lattice design. The experimental plot consisted of 4 rows, 2.2 m in length with 0.75 m and 0.2 m spacing between rows and plants, respectively. The gross and net plot sizes were 6.6 m2 (3 m × 2.2 m) and 3.3 m2 (1.5 m × 2.2 m), respectively. Seeds were sown by hand drilling at the rate of 12 kg ha-1 as per the recommendation for row planting in sorghum. Thinning was done two weeks after emergence to adjust plant pacing. The recommended NPS fertilizer was applied at the rate of 100 kg ha-1 during planting and Urea fertilizer was applied as a top dressing of 50 kg ha-1 at the knee height stage. The field was kept free of weeds by hand weeding during the whole growing period and other cultural practices were carried out as per standard practices recommended for the study area.
Data collection
Ten individual plants were selected randomly per plot and marked before panicle emergency and used as a sample for plant height (cm), leaf number per plant, leaf length, leaf width (cm), leaf area (cm2), panicle length (cm), head weight (g), panicle width (cm) and biomass yield (kg ha-1). Plot base data such as days to flowering, days to maturity, grain filling period, grain filling rate (kg/ha/day), stand count at harvest, thousand seed weight (g), grain yield (kg ha-1) and harvest index (%) were collected following the descriptors of sorghum [15].
Data analysis
Analysis of variance was computed for all traits as per the model for simple lattice design by using SAS Computer Statistical Package version 9.0. Means that show significant differences were compared using Duncan’s Multiple Range Test (DMRT) at a 5% significant level. The following model was used in the analysis of variance.
Pijk = µ + gi + rj+ + bk (j) + eijk
Where; Pijk = phenotypic value of ith genotype under jth replication and kth incomplete block within replication j; µ = grand mean; gi = the effect of ith genotype; rj = the effect of replication j; bk (j) = the effect of incomplete block k within replication j and eijk = the residual or effect of random error.
Estimation of coefficients of variation: The phenotypic and genotypic variations were computed using the formula suggested by [16] as follows.
Phenotypic variance (σ²p) = σ²g + σ²e
Where: Msg = mean square due to genotypes, Mse = error mean square, r = the number of replication, k = block size, σ²g = genotypic variance, σ²e = environmental variance, and σ²p = phenotypic variance.
Phenotypic and genotypic coefficients of variations were calculated according to the formula outlined by [17].
Where: PCV (%) = Percentage of phenotypic coefficient of variation
PCV (%) = Percentage of genotypic coefficient of variation
= Mean of the population for the trait.
The PCV and GCV values were categorized as 0% - 10% = low; 10% – 20% = moderate and > 20% = high values as indicated by [18].
Estimation of broad sense heritability: Heritability (H2) in a broad sense for all characters was computed using the formula adopted by [19].
Where; H2= heritability in a broad sense; σ²g = genotypic variance, and σ²p = phenotypic variance. The heritability percentage was categorized as low = 0 - 30%, moderate = 30 – 60, and high = > 60 suggested by [20].
Estimation of expected genetic advance: Genetic advance under selection (GA) for each character was computed using the formula adopted by [21].
and
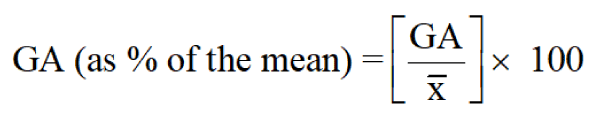
Where; GA = Genetic advance, k = selection differential (at 5% selection intensity with value 2.063), σp = phenotypic standard deviation, H2 = heritability, Grand mean. The GA as a percentage of the mean was categorized as low (0 - 10%), moderate (10-20%), and high (>20%) as suggested by [21].
Results and discussion
Analysis of variance
The result of the analysis of variance for 17 traits is presented in Table 2. The genotypes exhibited highly significant (p < 0.01) differences for all traits. The observed significant differences among genotypes for all traits indicated the presence of variability for each of the characters among the tested sorghum genotypes and a good opportunity for the breeders since it allows them to develop varieties of interest through selection and/or hybridization. The relative efficiency of simple lattice design was greater than one for most of the quantitative traits except panicle width, head weight, stand count at harvest, and thousand seed weight indicating a simple lattice design had an advantage over Randomized Complete Block Design (RCBD). The coefficients of variation were in the range of 2.34% for days to maturity and 14.05% for panicle width. The results of the relative efficiency of designs and coefficients of variation indicated the lattice design was efficient and reliable data were collected [22]. Several authors reported highly significant (p < 0.01) differences for quantitative traits among sorghum genotypes for days to flowering, days to maturity, plant height, thousand seed weights, head weight, grain filling period, grain filling rate, and grain yield similar to the present study findings [23-26].
Mean performance of genotypes
Phenology and growth traits: The variation for days to flowering was in the range between 66 and 79 days with a mean of 71.45 days. The late flowering was recorded for G8 (79 days) followed by G6 (78.5 days), G5 (78 days), G4 (77 days), and G59 (77 days), whereas the early flowering was recorded for G9 (66 days) followed by G56 (66.5 days), G41 (66.5 days), G54 (67 days) and G49 (67 days). Thirty-two genotypes were below the grand mean of days to flowering. Days to maturity ranged from 100 to 124 days with a mean of 112.67 days. The latest maturity date was recorded for Melkam (124 days) followed by G59 and G41 (123.5 days), and G16 (122 days). The early maturing genotypes were G49 (100 days), G47 (102 days), G44 (102.5 days), G36 (104.5 days), G45 (105.5 days), G23 (105.5 days) and G22 (106.5 days) and these genotypes were not significantly different from each other in maturing date (Table 3).
Early flowering and maturity are well-known drought escape mechanisms. Those early flowering and maturing genotypes would be appropriate for moisture stress areas while those genotypes with late flowering and maturity could be recommended for optimum moisture areas. Similarly, many authors reported a range of variation among sorghum landraces for days to flowering and maturity depending on the sorghum genotypes used and the locations where the genotypes were evaluated [12,27-29].
The genotypes showed 32.5 to 56 days for the grain filling period with a mean of 41.69 days and the grain filling rate ranged from 44.87 to 119.45 kg ha-1 days-1 with a of mean 77.31 kg ha-1 days-1. The result showed the presence of a wide range of variation among the genotypes for grain filling period and grain filling rate. This result is in line with [25,26] reported a wide range of variation among sorghum genotypes for grain filling rate of 41.7 to 191.4 kg ha-1 day-1. [30] also reported similar results for the grain-filling period that ranged between 34 and 47.50 days with a mean 41.94 of days.
The variation of genotypes for plant height ranged from 159.83 to 275.5 cm with a mean of 195.61 cm. Genotypes G56 (275.5 cm) followed by G14 (233.33 cm) and G29 (233 cm) were the tallest, whereas G1 (159.83 cm), G22 (160.34 cm) and G23 (160.84 cm) were the shortest genotypes. Most of the advanced genotypes were taller than the standard checks. The genotypes which showed tall plant height also produced higher biomass yield production in the study area (Table 3) [31] reported that tall genotypes are important genetic resources for fodder production and for house construction and as a thatching material in Ethiopia. On the other hand, the presence of variable plant height would be important for the selection of genotypes that fit a different purpose. For instance, short plant height and maturity have been identified as important traits for drought tolerance [32].
The overall average leaf number per plant was 7.95 ranging from 6.74 to 9.04. The highest number of leaves per plant (9.04) was produced by G37 followed by G58 (9.00), G29 (8.94), G59 (8.77), and G50 (8.77). The lowest number of leaves per plant was recorded from G49 (6.74), G44 (6.84), and G14 (6.87). Out of the evaluated genotypes, about 33 of the genotypes showed the highest leaf number per plant than the overall mean. Leaf length varied between 81.17 cm for two genotypes (G28 and G29) and 65.34 cm for G20 with an overall mean value of 74.07 cm. A total of 32 and 15 genotypes showed the highest leaf length than the overall mean and standard check (Erer), respectively. The observed variation indicated the opportunity to select genotypes with maximum leaf number and leaf length. The genotypes which had high leaf numbers per plant and leaf length might contribute to higher biomass yield production. This result is in line with [33] who reported a wide range of variation among 64 sorghum landraces for leaf number per plant that ranged from 9 to 15.66 and leaf length that ranged from 38 to 95 cm.
Leaf width ranged from 6.18 to 9.56 cm. The two genotypes G10 and G51 showed a maximum leaf width (9.56 cm), whereas a minimum leaf width was obtained from G46 (6.18 cm), G40 (6.37 cm), and G31 (6.48 cm). Forty genotypes exceeded the overall mean (8.14 cm) value of the tested genotypes in leaf width. The mean leaf area ranged from 224.39 to 528.39 cm2 with a grand mean of 443.81 cm2. Maximum leaf area was recorded from genotypes G10 (528.39 cm2) followed by genotype G16 (521.28 cm2), G51 (515.13 cm2), and G8 (514.54 cm2) while the lowest leaf area was recorded from genotype G55 (224.39 cm2). Similarly, [29] reported variation among sorghum genotypes for leaf areas between 351.57 and 390.80 cm2.
Yield and yield components: Wide ranges were recorded for yield and yield components (panicle length, panicle width, panicle weight, stand count at harvest, thousand seed weight, biomass yield, and grain yield) and harvest index (Table 4). Panicle length ranged from 20.17 cm for G35 to 31 cm for G17 with a mean value of 25.47 cm. Among genotypes, 63.33% showed superiority for panicle length over the mean value of genotypes. The mean value of panicle width was 12.69 cm with a maximum of 24 cm for G4 and a minimum of 7.5 cm for the Melkam variety. A total of 26 genotypes revealed superiority for panicle width above the overall genotypes mean (Table 4). This result is in line with [34] who evaluated 117 sorghum accession and reported a range between 12 and 36.4 cm for panicle length and 12 and 36.4 cm for panicle width.
The lowest panicle weight was obtained from G22 (85 g) while the highest was obtained from G4 (230 g) with a mean value of 128.73 g. Twenty-seven and twenty genotypes had higher panicle weight than the mean and best checks (Erer and Argiti) of 64 sorghum genotypes. Stand count at harvest was in the range of 28.5 for G34 to 40 for G3 with a mean value of 35.1. The mean thousand seed weight was 29.15 g with a maximum of 35 g and a minimum of 20.5 Thirty-two genotypes exceeded the overall mean value (29.15 g) of the tested genotypes, while 22 genotypes showed superiority over the standard check varieties (Melkam and Erer). Similarly, Kassahun, et al. (2011) reported thousand kernel weights that varied from 10.8 to 54.0 g. Wide ranges were recorded for panicle weight between 41.1 and 135.34 g with a mean value of 95.37 g and for thousand kernel weights between 45.58 and 20.60 g with a mean value of 33.41 g [35].
The maximum biomass yield was obtained from G17 (31000 kg ha-1) while the minimum biomass yield was obtained from G35 (20170 kg ha-1). For this trait, 40 and 22 of 64 genotypes had higher biomass yield than the mean biomass yield (25465 kg ha-1) and best check (Argiti). Therefore, there is plenty of variability among the genotypes for a selection designed for the improvement of this trait. In line with [36] the current study reported a higher range of variability in the biomass yield of sorghum genotypes.
Grain yield ranged from 2100 to 5305 kg ha-1 with a mean yield of 3308.83 kg ha-1. The highest grain yield was obtained from G50 (5305 kg ha-1) followed by 15 genotypes that gave grain yield in the range between 4500 and 3640 kg ha-1 with non-significance difference among them, while G42 (2100 kg ha-1), G58 (2110 kg ha-1), G2 (2190 kg ha-1) and G23 (2300 kg ha-1) had lower grain yields. Out of the evaluated sorghum genotypes, about 37 and 16 genotypes showed the highest grain yield over the grand mean and standard check (Melkam), respectively. Similarly, [25] reported grain yield ranging between 2085 and 3655.7 kg ha-1 with mean values of 3035 kg ha-1.
The harvest index for genotypes ranged from 8.12% for G57 to 20.21% for G50 with a mean of 12.70%. For this trait, about 37 and 4 genotypes showed superiority for harvest index over the overall mean and the standard check (Erer), respectively. In most cases, the improvement of the harvest index had been a consequence of increased grain yield relative to the increased biomass. Enhancing the harvest index increases the economic portion of the plant [37]. The harvest index is considered a measure of biological success in partitioning assimilated photosynthetic to the harvestable product [38]. Similarly [39,40], reported a harvest index which ranged from 3.73% to 24.55% of which 44 sorghum lines out of 100 exceeded the overall mean.
Estimation of variance components
Estimation of phenotypic variance (σ²p), genotypic variance (σ²g) and Phenotypic Coefficient of Variation (PCV), and Genotypic Coefficient of Variation (GCV) are provided in Table 5. The PCV ranged from 4.74% for days to flowering to 24.74% for panicle width, while GCV ranged from 3.58% for leaf length to 20.33% for panicle width. In general, the PCV values were greater than the GCV values for all characters, indicating the influence of the environmental effects and uncontrolled experimental error on the expression of these traits. High values of PCV and GCV were observed for panicle width, head weight, and harvest index. These high values of PCV and GCV revealed that the genotypes have a broad base genetic background and the existence of substantial variability to facilitate improvement through selection. Similarly [14], reported high PCV and GCV values for panicle width, head weight, and harvest index.
Traits with moderate PCV and GCV values were obtained for grain filling period, grain filling rate, plant height, panicle length, and grain yield, indicating substantial improvement could be obtained through recurrent selection for these traits. In addition, moderate PCV value was obtained for leaf width, leaf area, and thousand seed weight. This suggested that these traits were highly influenced by environmental factors and selection of genotypes based on the phenotypic mean values might not be rewarding [40]. Similarly, [41] reported moderate PCV for grain filling period, plant height, thousand seed weight, leaf width, and leaf area, and moderate GCV for grain filling period, plant height, panicle length, panicle width, and grain yield. Also [25] reported moderate PCV and GCV values in grain filling period, grain filling rate, plant height, panicle length, thousand seed weight, and grain yield.
Low values of PCV and GCV were estimated for days to flowering, days to maturity, leaf number per plant, leaf length, biomass yield and stand count at harvest. In addition, other traits such as leaf width, leaf area, and thousand seed weight had low GCV values. This showed that those traits were more influenced by environmental factors for their phenotypic expression and relatively smaller variability. Similarly, low PCV and GCV were reported for days to flowering, days to maturity, and leaf length [41-43] in previous studies.
Estimates of heritability and expected genetic advance: The estimated broad sense heritability and expected genetic advance are presented in Table 5. The heritability values for the 17 traits ranged from 24.74 for biomass yield to 96.6% for head weight. High heritability was estimated for most traits such as days to flowering, days to maturity, grain filling rate, plant height, head weight, panicle width, panicle length, grain yield, and harvest index. This result suggested that selection could be fairly easy and improvement is possible using these traits as selection criteria in sorghum breeding programs. The heritability of these traits is due to additive gene effects and selection may be effective in early generations for these traits [44]. This result is in agreement with [35] who reported high heritability estimates for days to flowering, days to maturity, plant height, panicle width, panicle length, grain yield, and harvest index in 100 sorghum genotypes [25] also reported high heritability for days to flowering, days to maturity, grain filling rate, plant height, head weight, panicle width, panicle length, and grain yield in sorghum genotypes.
Moderate heritability was observed for the grain filling period, leaf number per plant, leaf width, leaf area, stand count at harvest, and thousand seed weight. [45] suggested that selection should be delayed until the generations become more advanced for traits with moderate heritability. Low heritability was estimated for biomass yield. This implies that selection may be considered difficult or virtually impractical due to the masking effect of the environment on this trait.
Estimates of genetic advance as a percent of the mean (GAM) at 5% selection intensity ranged from 4.8% for biomass yield to 41.95% for panicle width (Table 5). High GAM was observed for grain filling rate, panicle length, panicle width, head weight, harvest index, and grain yield indicating the predominance of additive gene action and high potential for improvement of the traits under selection. In previous studies, high genetic advance as a percent of the mean for plant height, grain yield, grain filling rate, panicle weight, panicle length, and grain yield were obtained [25]. Also, similar results, high estimated values of genetic advance, expressed as a percent of the mean, for panicle width and harvest index have been reported by [46].
Moderate GAM was calculated for grain filling period, plant height, leaf width, leaf area, and thousand seed weight indicating that the traits were less influenced by environmental factors but governed by both additive and non-additive gene actions. Hence, a simple selection is suggested for further improvement in the later generations. Low GAM was estimated for days to flowering, days to maturity, leaf number per plant, leaf length, biomass yield and stand count at harvest. This implies that the improvement of these traits in genotypic value for the new population compared with the base population under one cycle of selection is <10% at 5% selection intensity. Low GAM observed for these traits indicated that the traits were under high environmental influence, and that selection based on these traits would be ineffective. The high broad sense heritability and low GAM of days to flowering, days to maturity, and leaf length may be due to the presence of a non-additive type of gene action [47].
High broad sense heritability alone does not always provide a high prediction of genetic gain to ensure effective selection for improvement; rather higher heritability coupled with a higher estimate of GAM [21]. Consequently, high heritability coupled with high GAM was observed in the case of grain filling rate, panicle width, panicle length, head weight, grain yield, and harvest index. This implies it could be very effective in improving upon selection. Similar results have been reported by [48].
Conclusion
The genotypes exhibited highly significant differences for all traits, indicating the presence of variability for each of the characters among the evaluated sorghum genotypes and a good opportunity for the breeders to develop varieties of interest through selection or hybridization. High H2 coupled with high GAM estimates was observed in the case of grain filling rate, panicle width, panicle length, head weight, grain yield, and harvest indicating that selection on phenotypic expression for these traits would be effective. Generally, the overall study revealed the presence of wide variability among the 64 sorghum genotypes evaluated which can be exploited to develop high-yielding varieties with desirable grain yield and early maturity in the study area where moisture stress is a critical problem for sorghum production. Based on the current results, genotypes such as G50, G32, G58, G3, and G37 were identified as having superior yield whereas G49, G47, G44, G36, and G23 genotypes were identified for early maturing. Therefore, the information generated from the current study can be used in sorghum breeding strategy for developing high-yielding and early maturing sorghum varieties.
I am also very grateful to Oromia Agricultural Research Institute (OARI) for financial support for the research work. My special thanks go to Fedis Agricultural Research Center (FARC) for providing me with the required facilities to conduct my research. I wish to thank all my friends of mine for creating an atmosphere of good working environment. I am also most grateful to Abdella Mustefa and Girma Wagari who supported me during data collection. My heartfelt thanks go to W/rt Abrat Girma and Kimiya Juneydi for closely managing my research work from planting to threshing.
- Chaudhary DP, Saini RK, Maurya BK, Sharma M, Kumar R, Sen R., Singh SK. Study of genetic variability and fodder yield components in forage sorghum (Sorghum bicolor L. Moench). Bulletin of Environment, Pharmacology and Life Sciences. 2018. 7(2): 05-09.
- Assefa S, Shimelis H, Mwadzingeni L, Laing M. Agromorphological characterisation and selection of sorghum landraces. Acta Agriculturae Scandinavica, Section B — Soil and Plant Science. 2018; 68(7): 585–595.
- Anagholi A, Kashiri A, Mokhtarpoor H. The study of comparison between inside forage sorghum cultivars and speed feed hybrids. Agricultural Science and Natural Resources Journal. 2000; 7(4): 73-83.
- Olani G, Feyissa T, Disasa T. Genetic diversity analysis of sorghum [Sorghum bicolor (L.) Moench] races in Ethiopia using simple sequence repeats (SSR) markers. African Journal of Biotechnology. 2021; 20(11): 431-439.
- FAO (Food and Agriculture Organization). 2021. Food and agriculture data. Available at: http://www.fao.org/faostat/en/#data/QC. (Accessed 14 April 2021).
- CSA (Central Statistical Agency). Agricultural Sample Survey: Report on area and production of major crops, Addis Ababa, Ethiopia. 2021.
- Adugna A. Assessment of yield stability in sorghum. African Crop Science Journal. 2007; 15(2): 83-92.
- Belay F, Meresa H. Performance evaluation of sorghum (Sorghum bicolor (L.) Moench) hybrids in the moisture stress conditions of Abergele District, Northern Ethiopia. Journal of Cereals and Oilseeds. 2017; 8(4): 26-32.
- Aditya JP, Bhartiya P, Bhartiya A. Genetic variability, heritability and character association for yield and component characters in soybean (G. max (L.) Merrill). Journal of Central European Agriculture. 2011; 12(1):27-34.
- Allard RW. Principles of Plant Breeding. 2nd Ed. John Willey and Sons, New York. 2000. 254.
- Amelework B, Shimelis H, Tongoona P, Laing M, Mengistu F. Genetic diversity of lowland sorghum landraces assessed by morphological and microsatellite markers. Australian Journal of Crop Science. 2016; 10(3): 291-298.
- Desmae H, David RJ, Ian DG. Geographic patterns of phenotypic diversity in sorghum (Sorghum bicolor (L.) Moench) landraces from North Eastern Ethiopia. African Journal of Agricultural Research. 2016; 11(33): 3111-3122.
- Lemma C. Genetic variability and character association among yield and yield related traits in lowland sorghum (Sorghum bicolor (L.) Moench) genotype, MSc Thesis, Haramaya University, Haramaya, Ethiopia. 2019.
- Mengistu G, Shimelis H, Laing M, Lule D, Mathew I. Genetic variability among Ethiopian sorghum landrace accessions for major agro-morphological traits and anthracnose resistance. Euphytica. 2020; 216(7): 1-15.
- IBPGR. Descriptors for sorghum [Sorghum bicolor (L.) Moench]. International Board for Plant Genetic Resources, Rome, Italy. 1993.
- Burton GW, De Vane E. Estimating heritability in tall fescue (Festuca arundinacea). Agronomy Journal. 1953; 45(7): 478-481.
- Singh RK, Chaudhary BD. Biometrical methods in quantitative genetic analysis. Biometrical methods in quantitative genetic analysis. 1977.
- Sivasubramaniah S, Meron M. Heterosis and in breeding depression in rice. Madras Agriculture Journal. 1973; 60:1139-1144.
- Allard R. Principles of plant breeding. John Wiley and Sons Inc. Publishing Co. Pvt. Ltd. New York, USA: 1960; 485.
- Robinson HF, Comstock RE, Harvey PH. Estimates of heritability and the degree of dominance in maize. Agronomy of Journal. 1955; 4: 353-359.
- Johnson HW, Robinson HF, Comstock RE. Estimates of genetic and environmental variability in soybeans. Agronomy Journal. 1955; 47(7): 314-318.
- Gomez KA, Gomez AA. Statistical Procedures for Agricultural Research. 2nd ed. John Wiley and Sons. Inc., New York, USA. 1984; 95-109.
- Gebreyohannes A, Tadesse T, Seyoum A, Nida H, Nega A, Senbetay T, Endalemaw C. Genetic variability in agronomic traits and associations in sorghum [(Sorghum bicolor (L.) Moench)] genotypes at intermediate agro-ecology sorghum growing areas of Ethiopia. African Journal of Agricultural Research. 2018; 13(49): 2780-2787.
- Haftu G, Mekbib F, Tadesse T, Lakew B, Ramesh PS. Correlation and path analysis of yield, yield contributing and malt quality traits of Ethiopian sorghum (Sorghum bicolor (L.) Moench) genotypes. African Journal of Plant Science. 2019; 13(8): 209-220.
- Endalamaw C, Semahegn Z. Genetic Variability and Yield Performance of Sorghum. International Journal of Advanced Biological and Biomedical Research. 2020; 8(2): 193-213.
- Senbetay T, Belete T. Genetic variability, heritability, genetic advance and trait associations in selected sorghum (Sorghum bicolor L. Moench) accessions in Ethiopia. Journal of Biology, Agriculture and Healthcare. 2020; 10(12): 1-8.
- Amare K, Zelleke H, Bultessa G. Variability for yield, yield related traits and association among traits of sorghum (Sorghum Bicolor (L.) Moench) varieties in Wollo, Ethiopia. Journal of Plant Breeding and Crop Science. 2015; 7(5): 125-133.
- Sabiel SA, Noureldin I, Baloch SK, Baloch SU, Bashir W. Genetic variability and estimates of heritability in sorghum (Sorghum bicolor L.) genotype grown in a semiarid zone of Sudan. Archives of Agronomy and Soil Science. 2016; 62(1): 139-145.
- Girma F, Mekbib F, Tadesse T. Temesgen Menamo and Kassahun Bantte. Phenotyping sorghum [Sorghum bicolor (L.) Moench] for drought tolerance with special emphasis to root angle. African Journal of Agricultural Research. 2020; 16(8): 1213-1222.
- Yali W. Genetic variability, heritability and associations of yield and its component traits among sorghum (Sorghum bicolor (L) Moench) lines in Miesso district, eastern Ethiopia. MSc Thesis, Ambo University, Guder, Ethiopia. 2021.
- Venkateswaran K, Elangovan M, Sivaraj N. Origin, domestication and diffusion of Sorghum bicolor. In Breeding Sorghum for diverse end uses. Wood head Publishing. 2019; 15-31.
- Birhan T, Bantte K, Paterson A, Getenet M, Gabizew A. Evaluation and genetic analysis of a segregating sorghum population under moisture stress conditions. Journal of Crop Science and Biotechnology. 2020; 23(1): 29-38.
- Warkad YN, Potdukhe NR, Dethe AM, Kahate PA, Kotgire RR. Genetic variability, heritability and genetic advance for quantitative traits in sorghum germplasm. Agricultural Science Digest. 2008; 28(3):165-169.
- Tesfaye K. Genetic diversity study of sorghum (Sorghum bicolor (L.) Moench) genotypes, Ethiopia. Acta Universitatis Sapientiae, Agriculture and Environment. 2017; 9(1): 44-54.
- Abebe E. Agro-morphological and nutritional quality evaluation of sorghum [Sorghum bicolor (L.) Moench] inbred lines in Sheraro, Northern Ethiopia. MSc. Thesis Haramaya University, Haramaya, Ethiopia. 2018.
- Gebregergs G, Mekbib F. Estimation of genetic variability, heritability, and genetic advance in advanced lines for grain yield and yield components of sorghum [Sorghum bicolor (L.) Moench] at Humera, Western Tigray, Ethiopia. Cogent Food and Agriculture. 2020; 6(1): 1764181.
- Li X, Yan W, Agrama H, Jia L, Jackson A, Moldenhauer K, Wu D. Unraveling the complex trait of harvest index with association mapping in rice (Oryza sativa L.). Plos Genetics Collection. 2012; 7(1): 29350.
- Furbank RT, Jimenez-Berni JA, George-Jaeggli B, Potgieter AB, Deery DM. Field crop phenomics: enabling breeding for radiation use efficiency and biomass in cereal crops. New Phytol. 2019 Sep;223(4):1714-1727. doi: 10.1111/nph.15817. Epub 2019 Apr 26. PMID: 30937909.
- Rakesh DC. Character association and genetic divergence studies in Sorghum bicolor Moench. MSc Thesis. Gwalior College of Agriculture. Gwalior (M.P), India. 2012.
- Upadhyaya HD, Wang YH, Sastry DV, Dwivedi SL, Prasad PV, Burrell AM, Klein RR, Morris GP, Klein PE. Association mapping of germinability and seedling vigor in sorghum under controlled low-temperature conditions. Genome. 2016 Feb;59(2):137-45. doi: 10.1139/gen-2015-0122. Epub 2015 Nov 24. PMID: 26758024.
- Aragaw G. Distribution, characterization and management of sorghum anthracnose (Colletotrichum sublineolum) in Eastern Ethiopia. Doctoral dissertation, Haramaya University, Haramaya, Ethiopia. 2020.
- Khadakabhavi S, Girish G, Yashoda Y. Character association and path analysis studies in germplasm lines of rabi sorghum (Sorghum bicolor (L.) Moench). Journal of Applied and Natural Science. 2017; 9(1): 206-210.
- Prasad BV, Sridhar V. Studies on genetic variability, correlation and path analysis in yellow pericarp sorghum [Sorghum bicolor (L.) Moench] genotypes. International Journal of Current Microbiology and Applied Sciences. 2019; 8: 367-373.
- Ali MA, Abbas A, Niaz S, Zulkiffal M, Ali S. Morpho-physiological criteria for drought tolerance in sorghum [(Sorghum bicolor (L.) Moench)] at seedling and post-anthesis stages. International Journal of Agriculture and Biology. 2009; 11(6): 674-680.
- Singh BD. Organisation for crop improvement in India. Plant Breeding: Principles and Methods. Kalyani Publishers, Ludhiana, 2001; 801-830.
- Mahajan RC, Wadikar PB, Pole SP, Dhuppe MV. Variability, correlation, and path analysis studies in sorghum. Research Journal of Agricultural Sciences. 2011; 2(1): 101-103.
- Ali ML, Rajewski JF, Baenziger PS, Gill KS, Eskridge KM, Dweikat L. Assessment .of genetic diversity and relationship among a collection of US sweet sorghum germplasm by SSR markers. Molecular Breeding. 2007; 10: 1007-1032.
- Goswami SJ, Patel PT, Gami RA, Patel RN, Khatri AB. Correlation and path analysis study of different characters for grain yield and fodder purpose in sorghum [Sorghum bicolor (L.) Moench]. Electronic Journal of Plant Breeding. 2020; 11(04): 1053-1061.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
