International Journal of Agricultural Science and Food Technology
Cluster and principal component analysis of maize inbred lines in low moisture stress areas in Ethiopia
Talefe Wendwessen*
Cite this as
Wendwessen T (2023) Cluster and principal component analysis of maize inbred lines in low moisture stress areas in Ethiopia. Int J Agric Sc Food Technol 9(3): 059-063. DOI: 10.17352/2455-815X.000194Copyright License
© 2023 Wendwessen T. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.This study used to cluster and principal component analysis to assess the agronomic and physiological variability of 40 maize inbred lines under low moisture stress conditions in Ethiopia. The study was conducted at Melkassa locations during the 2017 main season. The analysis of variance showed the mean square due to genotype was highly significant (p < 0.01) for all traits which indicates the existence of sufficient genetic variability and potential for selection and improvement on the characters. Cluster analysis revealed four distinct groups of inbred lines, The members of clusters 1 and 4 may be combined in future breeding programs to obtain genotypes/hybrids that can perform well under drought stress conditions. The inter-cluster D2 values varied from 48.65 to 4407 indicating the high range of diversity present among the genotypes. while principal component analysis showed the first six PCs having Eigen value >1 explaining principal components that accounted for 77.7% of the total variation among the inbred lines. The first three principal components PC1, PC2, and PC3 with values of 25.25%, 18.03, and 11.7% respectively more contributed. Grain yield, plant height, ear height, and the number of leaves per plant were the most important traits influencing the variability among the inbred lines.
Introduction
Maize (Zea mays L.) is one of the most important staple crops in Ethiopia, serving as a vital source of food security and livelihood for millions of people [1]. However, the productivity of maize is significantly affected by various abiotic stresses, including low moisture conditions [2]. In Ethiopia, where the majority of maize cultivation is rainfed, low moisture stress poses a major challenge to maize production in several regions [3].
Understanding the genetic diversity and population structure of maize inbred lines under low moisture stress conditions is crucial for the development of improved varieties with enhanced drought tolerance [4]. Cluster analysis and Principal Component Analysis (PCA) are powerful tools that can aid in unraveling the genetic variation and relationships among different maize inbred lines.
Cluster analysis allows the grouping of similar maize inbred lines based on their genetic characteristics, thereby facilitating the identification of distinct clusters or subgroups within the population [5]. This information can guide breeders in selecting parental lines for hybridization programs and identifying potential sources of drought tolerance.
On the other hand, PCA is a statistical technique that reduces the dimensionality of a dataset while preserving most of its variation. The most important factors or principal components that account for the majority of the genetic variance can be found by using PCA in the genetic data of maize inbred lines [6]. This approach enables a comprehensive understanding of the underlying genetic structure and relationships among the different inbred lines.
In this study, we aim to investigate the genetic diversity and population structure of maize inbred lines under low moisture stress areas in Ethiopia using cluster analysis and PCA. By employing these analytical approaches, we seek to identify genetically distinct clusters within the population and determine the key factors contributing to the genetic variation. The findings from this research will provide valuable insights for maize breeding programs, enabling the development of drought-tolerant maize varieties tailored to the specific low moisture stress conditions in Ethiopia.
Overall, this study aims to contribute to the advancement of maize breeding efforts in Ethiopia, enhancing the resilience and productivity of maize crops under low moisture stress. By understanding the genetic diversity and relationships among maize inbred lines, breeders can make informed decisions to develop improved varieties that address the challenges posed by low moisture stress, ultimately benefiting maize farmers and ensuring food security in Ethiopia.
Additionally, the findings of the paper have practical implications for maize breeding programs and agricultural practices in low moisture stress areas. By identifying genetically diverse maize inbred lines with desirable traits, the paper provides valuable guidance for the development of improved maize varieties that can withstand low moisture stress conditions. This information is crucial for enhancing crop productivity and ensuring food security in regions facing water scarcity.
It contributes to the broader field of agricultural research by expanding the knowledge base on maize breeding and adaptation to low moisture stress. It adds to the limited literature on the genetic diversity of maize inbred lines in Ethiopia and provides a valuable reference for future studies in similar environments. The paper’s insights can also stimulate further research and collaboration among scientists and breeders working on maize improvement in low moisture stress areas.
Materials and methods
The experiment was conducted during the 2017 main season in Ethiopia’s low moisture stress areas at Melkassa Agricultural Research Center (MARC), Central Rift Valley of Ethiopia, the center is located at an elevation of 1550 above sea level with the latitude of 8024’ N and longitude of 39021’ E. The average annual rainfall in the area is 768 mm, which is erratic and uneven in distribution. The site has a mean maximum temperature of 28.50C and a mean minimum temperature of 12.6 0C. Loam and clay loam soil textures are the dominant soils of the area [7].
Forty maize inbred lines were used in this study, selected from the Melkassa Agricultural Research Center (MARC) Maize breeding program. These inbred lines were chosen based on their tolerance to low moisture stress and their potential for yield improvement.
Experimental design and data collection
A Total of 40 inbred lines were obtained which were collected from Melkasa Agricultural Research Center (MARC) maize program and the lines were planted 8 x 5 alpha lattice design (Patterson and Williams, 1976) with two replications and two rows per plot. The plot length was four (4) meters and 75 x 25 cm spacing between rows and plants used, respectively. Thinning was done at the three to five leaf stages to attain a final plant density of 53,333 plants ha-1. All management practices including planting, fertilization, weeding, and harvest were as performed as per the recommendations for the location.
Collected data
Ten plants were selected randomly for recording observations of all the quantitative traits except for days to 50 % tasselling and silking. The mean of ten plants for each entry in each replication was worked out for each trait used to use for statistical analysis. The data recording was done for each quantitative trait (Days to 50% Anthesis, Days to 50% silking, Days to 90% Maturity, Plant height (cm), Ear height (cm), No. of leaves per plant, Leaf length (cm), Leaf width (cm), No. of nodes per plant, No. of tassel branches, Tassel length of the main branch, No. of ears per plant, Ear length (cm), Ear diameter (cm), No. of kernels per ro , No. of kernel rows per ear, 1000 kernel weight Grain yield (kg/plot), Grain Yield (kg/ha)).
Result and discussion
Principal component analysis
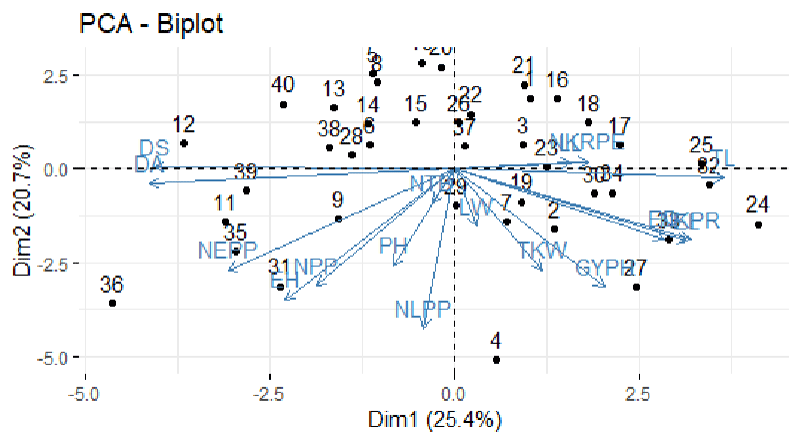
Principal Component Analysis analyzes a data table representing observations described by several dependent variables, which are, in general, inter-correlated. Its goal is to extract the important information from the data table and to express this information as a set of new orthogonal variables called principal components.
According to the principle of Syafii, et al. [8], the first principal component accounts for maximum variability in the data concerning succeeding components.
The principal component analysis showed that the gross variability observed among the 40 test genotypes can be explained with six principal components with eigenvalues greater than one as shown (Table 1). The first six components together accounted for about 77.7% of the total variation among the genotypes concerning all the 18 traits evaluated and showed the presence of considerable genetic diversity among the genotypes for most of the traits under consideration. Individually, PC1, PC2, PC3, PC4, PC5, and PC6 in that order accounted for about 25.25%,18.03% 11.7% 9.3% 7.9%,55% of the gross variation among the 40 maize genotypes evaluated for 18 traits.
The traits which contributed more to PC1, were node per plant, Ear length, days to Anthesis, days to silking, and Ear length. Whereas for PC2 number of kernels per row, number of leaves leaf per plant, ear diameter, plant height, and tassel length, For PC3, days to Anthesis, days to silking, 1000-kernel weight, Ear length, and Ear diameter, for PC4, the number of kernel row per Ear number of tassel branch, leaf length, leaf width for PC5, Leaf length, Ear height and days to Anthesis, days to silking, leaf width, Ear length and for PC6, days to maturity and leaf width. The first three principal components PC1 PC2 and PC3 with values of 25.25%, 18.03, and 11.7% respectively more contributed.
The present study confirmed that the Maize genotypes showed significant variations for the characters studied and it suggested many opportunities for genetic improvement through selection. Similar works have been done by [9], Dawit, et al. (2012) and [10,11] for grouping genotypes by principal component analysis (Figure 1).
Cluster analysis
Genetic divergence is the process in which two or more populations of an ancestral species accumulate independent genetic changes (mutations) through time, often after the populations have become reproductively isolated for some time. An appropriate number of clusters was determined by using points where local peaks of Pseudo F-statistics join with small values of the Pseudo t2 statistics followed by a larger Pseudo t2 for the next cluster fusion. Genetic distance between pairs of clusters was calculated using the Mahalanobis generalized distances were utilized to estimate the distances between and within clusters using the SAS computer software package as per the following formula: D2 ij = (Xi-Xj)’ S-1(Xi-Xj). where, D2 ij = the distance between any two groups i and j; Xi and Xj are the vector mean of the traits for the ith and jth groups, respectively, and S-1 = the inverse of the pooled covariance matrix . (Singh and Chaudhary,1996). The D2 statistics measure the force of differentiation at intra-cluster and inter-cluster levels (Singh, 2007).To assess the total variations and supplement the cluster analysis, principal component analysis was also carried out using the SAS computer software facilities (SAS, 2001), involving all 18 quantitative traits.
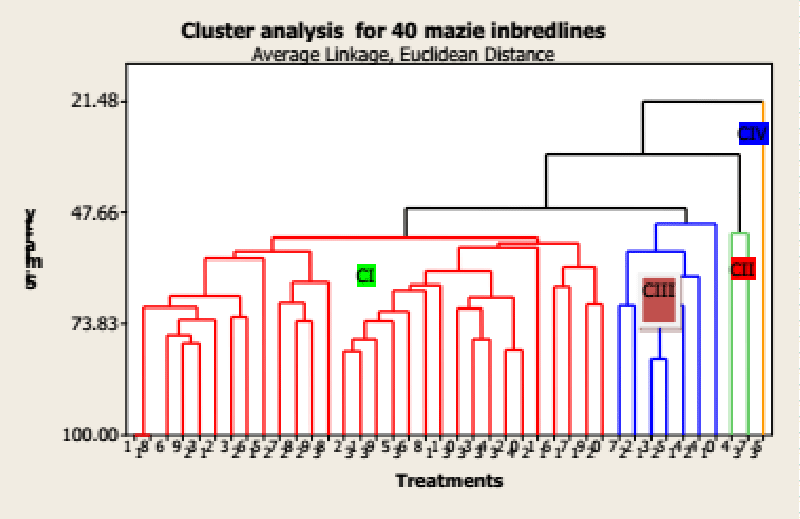
The intra and inter-cluster values among the four clusters are presented in Table 2. The inter-cluster D2 values varied from 48.65 to 4407 indicating the high range of diversity present among the genotypes. The highest inter-cluster divergence was noted between cluster I and IV followed by II and IV. It indicated that the genotypes grouped in these clusters were highly divergent from each other. Selection of parents from the highly divergent clusters is expected to manifest high heterosis in crossing and also wide variability in genetic architecture. Murty and Anand [12] claimed that there is a positive relationship between the specific combining ability and the degree of genetic diversity. The intra-cluster D2 values varied from zero (cluster I) to 48.64 (cluster III).
Clustering of genotypes
The main aim of using a clustering technique in the analysis of data from plant breeding trials is to group the varieties into several homogeneous groups such that those varieties within a group have a similar response pattern across the locations. It is reasonable to suppose that all the varieties in the trials will not behave completely independently of one another. For instance, those with similar genetic makeup would be expected to behave similarly. This study was conducted to provide information about the performance of maize lines in terms of morphological parameters, which will be helpful to maize breeders for further manipulation in subsequent breeding programs.
Forty genotypes were grouped into four clusters and the Euclidean distance matrix of 40 Maize genotypes estimated from 18 quantitative traits was used to construct Dendrograms based on the Unweighted Pair-group methods with Arithmetic Means (UPGMA) using Ward’s minimum variance procedure [13] and the distributions of the genotypes into different clusters are depicted in Table 3 and Figure 2. Accordingly, the 40 maize genotypes were grouped into four distinct clusters (Figure 2 and Table 2).
Cluster I was the largest and consisted of thirty genotypes Cluster II had Two genotypes, Cluster III Shad even genotypes, and Cluster IV had one genotype. This indicates that the crossing between superior genetic divergences of the above diverse clusters might provide desirable recombinants for developing high-yielding maize genotypes. This is because the cluster analysis sequestrates genotypes into clusters that exhibit high homogeneity within a cluster and high heterogeneity Between Clusters (Jaynes, et al. 200) [14,15]. In general, the intra-cluster distance was much less than the inter-cluster one. Similarly, Ahamed, et al. [16] reported that the inter-cluster distances were higher than the intra-cluster distances.
Cluster mean analysis
The mean values of four clusters which indicate their broad genetic bases for 18 quantitative characters are presented in Table 4. Cluster IV had mean values greater than the overall mean values of genotypes for days to flowering, days to silking, plant height, ear height, node per plant, number of kernels per row, and grain yield per hectare. In addition, Cluster III had mean values greater than the overall mean values of genotypes for Ear diameter, Number of Ears per plant, number of tassel branches, number of kernel rows per ear, and thousand kernel weights. Cluster II had mean values greater than the overall mean values of genotypes for plant height ear height, ear length Ear diameter, number of leaves per plant, leaf length, leaf width, tassel length, number of tassel branches, number of kernels per row, number of tassel branch and grain yield per hectare. Cluster I had mean values greater than the overall mean values of genotypes for numbers ear per plant, tassel length, and grain yield per hectare. These clusters had mean values lower than the overall mean values of genotypes for the remaining traits.
Singh, et al. (2014)[17-20] also reported that the character contributing the maximum to the divergence should be given greater emphasis for deciding the type of cluster for further selection and the choice of parents for hybridization.
Conclusion
The study on the cluster and principal component analysis of maize inbred lines in low moisture stress areas in Ethiopia aimed to identify distinct groups among the lines and explore relationships among variables. The findings provide insights into genetic diversity and potential drought tolerance of maize inbred lines in these regions. Cluster analysis categorized lines into groups, revealing diverse genetic resources for breeding programs. Principal Component Analysis (PCA) identified key variables influencing drought tolerance. Comparing cluster analysis and PCA results revealed relationships between genetic clusters and important drought-related traits. This study guides breeders in selecting suitable inbred lines for breeding programs, promoting genetically diverse clusters. The research contributes to developing resilient maize varieties, benefiting farmers and enhancing food security. Further research and utilization of identified clusters and influential traits are recommended for ongoing maize breeding efforts.
- Shiferaw B, Prasanna BM, Hellin J, Bänziger M. Crops that feed the world 6. Past successes and future challenges to the role played by maize in global food security. Food security. 2011;3:307-327.
- Jemal K, Berhanu S. Effect of Water Application Methods in Furrow Irrigation Along with Different Types of Mulches on Yield and Water Productivity of Maize (Zea Mays l.) at Hawassa, Ethiopia [dissertation]. Haramaya University; 2018.
- Tesfaye A, Yirga C, Alemu D, Bekele A. Situation and outlook of maize in Ethiopia.
- Vido J, Nalevanková P, Valach J, Šustek Z, Tadesse T. Drought analyses of the Horné Požitavie region (Slovakia) in the period 1966-2013. Adv Meteorol. 2019.
- Mengesha WA, Menkir A, Unakchukwu N, Meseka S, Farinola A, Girma G, Gedil M. Genetic diversity of tropical maize inbred lines combining resistance to Striga hermonthica with drought tolerance using SNP markers. Plant Breeding. 2017;136(3):338-343.
- Yang X, Gao S, Xu S, Zhang Z, Prasanna BM, Li L, Yan J. Characterization of a global germplasm collection and its potential utilization for analysis of complex quantitative traits in maize. Mol Breeding. 2011;28:511-526
- Ministry of Agriculture (MoA). Agro-Ecological Zones of Ethiopia, Natural Resource Management and Regulatory Department, Addis Ababa, Ethiopia. Draft document; 2000.
- Syafii M, Cartika I, Ruswandi D. Multivariate analysis of genetic diversity among some maize genotypes under maize-albizia cropping system in Indonesia. Asian J Crop Sci. 2015;7(4):244-255.
- Bhusal TN. Discrimination of maize (Zea mays L.) inbreeds for morpho-physiological and yield traits by D2 statistics and principal component analysis (PCA). Asian J Biosci. 2016;11(2):77-84.
- Vol AS. Cluster and principal component analyses of maize accessions under maize-albizia cropping system in Indonesia. J Agric Sci. 2015;60(1):33.
- Shazia G. Genetic Diversity in Maize (Zea Mays L.) Inbred Lines from Kashmir. Int J Pure App Biosci. 2017;5(1):229-235.
- Murty BR, Arunachalam V. The nature of genetic divergence about breeding systems in crop plants. Indian J Genet. 1966;26A:188-198.
- Anderberg MR. Cluster Analysis for Application. New York: Academic Press; 1993.
- Hafiz SBM, Jehanzeb F, Ejaz-ul-Hasan, Tahira B, Mahmood T. Cluster and principal component analyses of maize accessions under normal and water stress conditions. Asian J Crop Sci. 2015;7(4):244-255.
- Sandeep S, Bharathi M, Narsimha RV, Eshwari KB. Principal component analysis in inbreds of Maize (Zea Mays L.). Int J Trop Agric. 2015;33(2):213-216.
- Ahamed KU, Akhter B, Islam MR, Alam MA, Humauan MR. Assessment of genetic diversity in cowpea [Vigna unguiculata (L.) Walp.] germplasm. Bull Inst Trop Agric Kyushu Univ. 2014;37:57-63.
- Singh G, Kumar R, Jasmine. Genetic parameters and character association study for yield traits in maize (Zea Mays L.). J Pharmacogn Phytochem. 2017;6(5):808-813. Available from: http://www.phytojournal.com/archives/2017/vol6issue5/PartL/6-5-14-319.pdf.
- Sivasubramaniah S, Meron M. Heterosis and inbreeding depression in rice. Madras Agric J. 1973;60:1139.
- Solomon C. Phenotypic evaluation and molecular studies of parents and recombinant inbred lines of teff [Eragrostis tef (Zucc) Trotter] [dissertation]. Haramaya University; Ethiopia.
- Umar UU, et al. Studies on genetic variability in maize (Zea mays L.) under stress and non-stress environmental conditions. Int J Agron Agric Res. 2015;7(1):70-77.
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
