International Journal of Agricultural Science and Food Technology
Genetic variability, heritability, genetic advance and divergence in Ethiopian cowpea [Vigna unguiculata (L) Walp] landraces
Tigray Agricultural Research Institute (TARI), Abergelle Agricultural Research Center, P. O. Box 44, Abi-Adi, Ethiopia
Author and article information
Cite this as
Belay F, Fisseha K (2021) Genetic variability, heritability, genetic advance and divergence in Ethiopian cowpea [Vigna unguiculata (L) Walp] landraces. Int J Agric Sc Food Technol. 2021; 7(1): 138-146. Available from: 10.17352/2455-815X.000101
Copyright License
© 2021 Belay F, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Information on genetic variability in cowpea germplasm is important for crop improvement and for efficient utilization of the existing genetic resources. Hence, the objectives of the present investigation were to estimate genetic variability, heritability, genetic advance and to identify divergent parents from distantly related clusters among Ethiopian cowpea accessions. The field experiment was carried out using 42 accessions at Abergelle Agricultural Research Center on station during the 2019 cropping season. Data were collected for 8 agronomic traits and analysis of variance revealed significant differences (p<0.01) among the accessions for the traits studied. Seed yield had higher Genotypic Coefficient of Variation (GCV) and Phenotypic Coefficient of Variation (PCV) coupled with the highest genetic advance as percent of mean (100%). All the traits had moderate (68.01) to very high (99.98%) broad sense heritability. Further, high heritability coupled with high genetic advance as percent of mean was attained for days to flowering, grain filling period, plant height, pod length, seed yield and thousand seed weight reflecting the presence of additive gene action for the expression of these traits and improvement of these traits could be done through selection. The cluster analysis based on agronomic traits revealed four distinct groups at 90% similarity level. The highest inter cluster D2 was recorded between cluster III and cluster IV (D2=133.69 units). The range of inter cluster distance was 15.25 to 133.69 units, respectively. In conclusion, the high genetic distance revealed among clusters has to be exploited via crossing and selection of the most divergent parents for future cowpea improvement program.
Cowpea [Vigna unguiculata (L.) Walp., Fabaceae (2n = 2x = 22)] is an important dual purpose (food and forage) legume crop widely grown under low input production systems and in arid and semi-arid agro-ecologies of the world [1,2]. It is predominantly a self-pollinated crop, with natural cross-pollination up to one percent. Cowpea could play significant role in mitigating malnutrition such as micronutrient deficiencies for poor farmers of Sub-Saharan countries [3]. It has distinct features such as its earliness in maturity, tolerance to drought, heat, acidity and low fertility, and seed types with high protein content and low cooking time [4]. It is well adapted to the drier regions of the tropics, where other food legumes do not perform well. Cowpea is eaten in the form of dry seeds, green pods, green seeds and tender green leaves. As a pulse crop, cowpea provides more than half the plant protein for human diets in some areas of semi-arid tropics is being referred to as “poor-man’s meat” [5].
Cowpea grain typically contains 230-250 g/kg Crude Protein (CP) and 500-670 g/kg starch on a Dry Matter (DM) basis and cowpea forage, i.e. the crop residue after harvesting grain, 210g CP and 600g digestible dry matter per kg DM. These excellent nutritional qualities of cowpea make as one of the potential crop as a component of the cropping system and livelihood for the smallholder farmers living in drier regions of Ethiopia [6]. Thus, this crop can contribute greatly towards meeting the food requirement of people in areas where food security and malnutrition are major challenges. Regardless of the various merits of cowpea in Ethiopia, the national production and productivity is far below the potential due to several abiotic and biotic constraints among which drought, insect pests, parasitic weeds and virus facing cowpea production to have resulted in a very low yield [7].
Though, Ethiopia is one of the centers of origin and/or diversity of cowpea [8] and more than 66.5% of arable land is very suitable for cowpea production [9], the country has not been in a position to be benefited from international and continental cowpea improvement program or from the national pulse crops research. This is because low attention in research for cowpea is given as compared to other pulse crops. To harness the potential of cowpea landraces grown in Ethiopia baseline information regarding cowpea production status in the country /baseline information has been generated. Greater the variability in a population, there is the greater chance for effective selection for anticipated varieties. However, very few studies have been conducted on cowpea genetic variability using quantitative traits employed in the country. Hence, the objectives of the present study were to assess variability, heritability, genetic advance and to identify divergent parents from distantly related clusters for the future cowpea improvement program.
Materials and methods
Description of the experimental site
The field trial was conducted at Abergelle district central zone of Tigray, northern Ethiopia during the 2019 main cropping season. The site (Mearey) is located at about 903 km north of Addis Ababa and 120 km south west of Mekelle city. It has an approximate geographical coordinates of 13014’06” N latitude and 38058’50” E longitudes having an altitude 1560 meter above sea level (Figure 1). The area is characterized by an erratic rainfall pattern with “kola” agro climatic zone. The rainy season is mono modal pattern concentrated in one season during the summer (July to August) and receives from 350- 600 mm, annual precipitation. The mean minimum and maximum temperature of the area ranges from 18- 420C, respectively [10]. The soil texture of the specific site of the study area is sandy clay textural class with high available P (13.82 mg kg-1), very low in total N (0.08%) and low organic matter (0.72%) with a neutral pH of 7.18.
Description of the planting materials
The experimental plant materials comprised a total of 42 cowpea accessions/local landraces along with one released variety Bekur as a check was used in this study. The landraces were Ethiopian origin kindly provided by the Ethiopian Institute of Biodiversity (EIB) collected from different agro-ecological regions of the country, varying in altitude, rainfall, temperature, and soil type. The accession numbers and source of the genotypes are presented in Table 1.
Experimental design and crop management
The experiment was laid out using (6,7) α- lattice design with three replications. The plot size was 7.2 m2 (4m x 1.8m) with three rows of inter-row (60 cm) and intra-row (20 cm) spacing’s. The distance between plots, intra-blocks, and replications was 0.5m, 1m and 1.5m, respectively. Blended NPSZnB fertilizer was applied at the rate of 100 kg ha-1 during planting. Weeds were controlled periodically by hand weeding and other field management and crop protection activities were done as required.
Data collection
Some phenological (days to flowering, days to maturity, grain filling period), morphological (plant height, pod length) and yield and related traits (seed yield, thousand seed weight, number of seeds per pod) of each genotype was collected following the descriptor for cowpea developed by the International Board for Plant Genetic Resources [12]. The data collected on plot basis were days to flowering, days to maturity, grain filling period (days), thousand seed weight (g) and seed yield (g). In addition, the data collected on individual plant basis were plant height (cm), pod length (cm) and number of seeds per pod. For single plant based traits, the average of data from the five random samples of plants per plot were used and seed yield per plot in grams was converted to kg ha-1 for analyses.
Data analysis
Data for agronomic traits were subjected to analysis of variances (ANOVA) for lattice design procedures of SAS Version 9.2 [13] to test the presence of significant differences among genotypes. Variability among accessions was estimated using genotypic variances and coefficients of variations as suggested by Burton and De vane [14] and these components of variance (δ2p, δ2e, δ2g) were used for the estimation of coefficients of variation (PCV, GCV) as described by Singh and Chaudhary [15].
1. Genotypic Variance GV= , where MSg = Mean Square of genotypes, MSe = Mean Square of error (environmental variance or δ2e), and r = number of replications;
2. Phenotypic Variance, PV= GV +MSe, where GV = Genotypic Variance and MSe = Mean Square of error;
3. Genotypic Coefficient of Variation, GCV = , where GV = Genotypic Variance and x =grand mean of the character;
4. Phenotypic Coefficient of Variation, PCV = , where PV = Phenotypic Variance and x = mean of the character.
5. Broad sense heritability (H2) of all traits was calculated according to the formula as described by Allard [16] as follows: H2 = [(σ2g) / (σ2p)] × 100, where σ2g and σ2p are genotypic and phenotypic variances respectively.
6. Genetic Advance (GA) for selection intensity (K) at 5% was computed according to Allard [16] as given here: GA = Kσ2pH2, where, GA = expected genetic advance, K = the standardized selection differential at 5% selection intensity (K=2.063), σ2p = is phenotypic standard deviation on mean basis and H2 = heritability in broad sense.
7. Genetic advance as percentage of population means (GAM) was also estimated with the methods described by Johnson, et al. [17] to compare the extent of the predicted advance of different traits under selection using the following formula: GAM= , where GA = genetic advance under selection and x = mean of the population.
8. Hierarchical [18] cluster analysis was performed to group accessions and construct a dendrogram by Ward’s method by using SAS software. The measure of dissimilarity was Euclidean distance. The average inter-cluster distance was calculated using the generalized Mahalanobis’s D2 statistics [19]. The R2 (RSQ), Cubic Clustering Criteria (CCC), Pseudo-F Statistics (PSF) and pseudo-T2 statistics were considered for defining optimum cluster numbers [20]. The contributions of each of the traits to divergence were estimated as described as Sharma [21] with the formula [CTIC= ], where SD and x are the standard deviation and mean performance of each trait, respectively.
Results and discussion
Genetic variability
The mean squares and estimates of phenotypic (σ2p), genotypic (σ2g) and environmental (σ2e) variances, Phenotypic Coefficients of Variation (PCV) and Genotypic Coefficients of Variation (GCV) along with the mean of traits investigated are presented in Table 2. The analysis of variance showed that the mean squares for the genotypes were highly significant (p<0.01) for all agronomic traits indicating presence of adequate variability among Ethiopian cowpea accessions. Genetic variance ranged from 1.68 for number of seeds per pod to 590157.45 for seed yield (kg ha-1) while phenotypic variance values ranged from 2.47 to 590283.96 for seed yield (kg ha-1). The GCV ranged from 6.90% for days to maturity to 48.80% for seed yield. Similarly, PCV ranges from 7.45% for days to maturity to 48.81% for seed yield. The GCV and PCV values are normally categorized as low (<10%), moderate (10-20%) and high (>20%) as indicated by Deshmukh, et al. [22].
The highest Phenotypic Coefficient of Variation (PVC) and Genotypic Coefficient of Variation (GCV) were recorded for seed yield (48.81, 48.80) followed by 1000-seed weight (33.36, 33.37) and grain filling period (23.25, 21.15) while low PVCs and GVCs were recorded for days to maturity (7.45, 6.90), number of seeds pod-1 (13.53, 11.16), plant height (13.96, 13.88), days to flowering (15.01, 14.80) and pod length (17.82, 16.44), respectively. The present study suggests that the phenotypic coefficient of variation (PCV) was relatively higher than the corresponding Genotypic Coefficient of Variation (GCV). However, the difference between PCV and GCV were narrow indicating little influence of environment on the expression of these traits and considerable amount of variation was observed for the traits studied.
The results of the study was in conformity with the findings of Reshma, et al. [23] who reported high value of PCV and GCV for seed yield per plant, 100-seed weight and number of pods per plant. Additionally, Manggoel, et al. [24] and Mofokeng, et al. [25] also reported high PCV and GCV for days to flowering, number of branches, pod number per plant, pod weight per plant, seed number per pod, hundred seed weight and seed yield in cowpea.
Broad sense heritability
The estimate of broad sense heritability for all the traits under study is presented in Table 2. All agronomic traits recorded high heritability ranged from 68.01(number of seeds per pod) to 99.98% (seed yield) indicating additive gene effects control the expression of the traits. Heritability is classified as low (<40%), medium (40–59%), moderately high (60-79%) and very high (≥80%), according to Singh [26]. In the current study, the highest estimates of broad sense heritability were recorded for seed yield (99.98) followed by plant height (98.78), thousand seed weight (98.29), days to 50% flowering (97.17), days to maturity (85.57), pod length (85.11), grain filling period (82.79) and number of seeds per pod (68.01). Similarly, Manggoel, et al. [24], Thorat and Gadewar [27], Khan, et al. [28], Khanpara, et al. [29], Reshma, et al. [23] and Mofokeng, et al. [25] reported high heritability values in cowpea.
Expected genetic advance for selection
Genetic advance is a measure of predetermined progress under artificial selection program. According to Jonhson, et al. [17] the value of GAM is categorized as low (< 10%), moderate (10-20%) and high (> 20%). In this study, the highest GAM was recorded for seed yield (100%) followed by 100-seed weight (68.16%), grain filling period (39.65%), pod length (31.25%), days to flowering (30.05%) and plant height (28.42%), indicating that these traits are governed by additive genes and selection will be rewarding for improvement of cowpea for these traits. In agreement with the current study, high GAM for days to flowering, number of branches, pod number per plant, pod weight per plant, seed number per pod, hundred seed weight, seed number per plant and seed yield [25]; number of pods per plant and 100 seed weight [30]; pod weight, number of pods per plant, days to 50% flowering, and number of seeds per pod [29,30]; number of branches per plant, plant height, pod yield per plot, total number of pods per plant and number of seeds per pod [31] were reported in cowpea.
Furthermore, high heritability coupled with genetic advance were recorded for days to flowering, grain filling period, plant height, pod length, seed yield and thousand seed weight, indicates additive gene action control the expression inheritance of these traits in cowpea [32]. A similar result was reported by Thorat and Gadewar [27], Sharma, et al. [33] and Das, et al. [34]for seed yield and Khan, et al. [28] for the number of pods per plant and Reshma, et al. [23] for plant height, seed yield per plant, number of pods per plant, pod length and hundred seed weight in cowpea. Overall, the estimates of heritability (H2), genetic advance as percent of mean (GAM); genotypic coefficients of variation (GCV) and phenotypic coefficient of variation (PCV) were high for seed yield and thousand seed weight which are critical to identify potential for development of superior cowpea genotypes and/or improvement of population through selection.
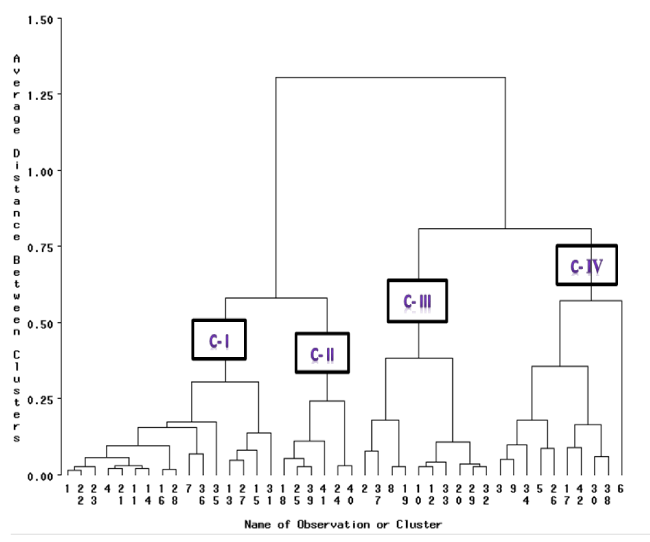
Clustering of accessions
The tested accessions were grouped in four different clusters with the number of accessions per cluster varying from 6 to 16 (Table 3). The covariance matrix gave hierarchical clustering (Figure 2) using average linkage method and the appropriate number of clusters was determined from the values of Pseudo F and Pseudo T2 statistics among 42 cowpea accessions (Table 4). Cluster I was the largest cluster comprising 16 accessions, followed by clusters II and IV that contained 10 and 10 accessions, respectively, where cluster III contained the smallest accession (6) number. Of the 16 accessions grouped in cluster I, 43.75%, 25%, 12.5%, 12.5% and 6.25% of accessions originated from Oromia, Tigray, Amhara, SNNP and Gambella regions, respectively.
Similarly, in cluster II 50%, 20%, 20% and 10%; cluster IV 40%, 30%, 20% and 10% of the total accessions grouped in the second largest clusters (Cluster II and IV) had in that order origins that are Gambella, B/Gumuz, SNNP and Oromia; Amhara, Oromia, Tigray and SNNP while the least of accessions (6) grouped in cluster III, 66.66%, 16.67% and 16.67% were originated from Amhara, Gambella and B/Gumuz regions of Ethiopia, respectively (Table 4). The cowpea accessions originating from the same regions entered into different clusters indicating the absence of relationships between genetic diversity and geographic origin. For instance, the accessions from Oromia, Amhara, SNNP, Tigray Gambella and B/Gumuz regions of origin grouped into the four distinct clusters (Tables 3,4).
The main reasons for the grouping of accessions of the same origin into different clusters could be the exchanges of germplasm by farmers among neighboring regions, natural and artificial selection, genetic enrichment, genetic drift and environmental variation. Furthermore, Tesfaye, et al. [35] reported no relationship between genetic origin and /or diversity and geographic distribution.
Mean performance of clusters
The mean value of 8 agronomic traits per cluster is presented on Table 5. In this study, the mean values varied among clusters for the traits studied. Accessions those took longer days to flowering, maturity, plant height and extended grain filling period were found in cluster III. Cluster IV exhibited maximum mean values for pod length, seed yield and thousand seed weight. On the contrary, no significant variation was recorded among clusters for number of seeds per pod.
On the basis of overall mean performance, cluster IV showed the best performance for most important traits including seed yield. Therefore, cluster IV would be preferable for selection of parents with high mean values for the improvement of accessions. Conversely, cluster III had minimum values for yield and yield related traits. It showed the poorest performance of traits while the highest plant height was recorded in this cluster. Therefore, this cluster is preferable for increasing number of pods per peduncle. In general, there was highly significant variation in mean performance among the clusters for most of the traits, and this offers a huge opportunity to select potential parents across the clusters for specific traits for future cowpea improvement program. Overall, the variation observed among the 42 cowpea accessions suggests that agronomic traits can reveal diversity existing among cowpea genotypes. Molosiwa, et al. [36], Moolendra, et al. [37] and Tesfaye, et al. [35] had also reported similar results.
Seed yield and thousand seed weight were the major contributors for genetic divergence to the entire accessions (Table 5) while days to flowering, grain filling period, and days to maturity, plant height, pod length and number of seeds per pod had small contribution towards genetic divergence. In agreement with the study; Tesfaye, et al. [35] classified the levels of trait contribution for inter cluster divergence for cowpea ≥15% as high contributor, ≥ 8% < 15% as medium contributor and <8% as little contributor for inter cluster divergence in Ethiopia [38-41].
Genetic divergence analysis
The standardized Mahalanobis D2 (square distances) statistics showed that there is high genetic distance and highly significant variation at P≤0.01 and P≤0.05 among the four clusters (Table 6), indicating wide diversity among cow pea accessions. The highest average inter-cluster distance was recorded between the cluster III and IV (D2=133.69 units) followed by the cluster I and IV (D2=69.39 units), cluster II and III (D2=49.86 units) while the lowest was between cluster I and II (D2=15.25 units). Overall, the present study indicates that the accessions in cluster III and V were more diverged than any one of the other clusters while the nearest inter-cluster distance between cluster I and II were not genetically diverse. Thus, the accessions belonging to the distant clusters could be used for cowpea breeding program to get a wider range of variability in the segregating F2 population.
Conclusion
The result of the study revealed the existence of significant (p<0.01) genetic variability among Ethiopian cowpea landraces. Seed yield had higher Genotypic Coefficient of Variation (GCV) and phenotypic coefficient of variation (PCV) coupled with the highest genetic advance as percent of mean (100%). Further, high heritability coupled with high genetic advance as percent of mean was recorded for days to flowering, grain filling period, plant height, pod length, seed yield and thousand seed weight reflecting the presence of additive gene action for the expression of these traits and improvement of these traits could be done through selection. The cluster analysis based on agronomic traits revealed four distinct groups at 90% similarity level. The highest inter cluster D2 was recorded between cluster III and cluster IV (D2=133.69 units). The range of inter cluster distance was 15.25 to 133.69 units, respectively. Therefore, based on the present findings, it can be conclude that the high genetic distance revealed among clusters has to be exploited via crossing and selection of the most divergent parents for future cowpea improvement program.
The authors thank Abergelle Agricultural Research Center for funding the research study. They also express their great gratitude to researchers of the center for devoting their time to effectively support this work.
- Timko MP, Singh BB (2008) Cowpea, a multifunctional legume. Genomics of Tropical Crop Plants 227–257. Link: http://bit.ly/2Pgw6Ln
- Srivastava S, Chopra Ak, Sharma P, Kumar V (2016) Amendment of sugar mill waste water irrigation on soil bio hydrological properties and yield of [Vigna umguiculata(L.) Walp.] in two seasons. Communication in Soil Science and Plant Analysis 48: 511-523. Link: https://bit.ly/2PcXTw4
- Menssena M, Marcus L, Emmanuel OO, Mary AO, Fekadu FD, et al.(2017) Genetic and morphological diversity of cowpea ( Vigna unguiculata (L.) Walp.) entries from East Africa. Scientia Horticulturae 226: 268-276. Link: https://bit.ly/2ON2dm3
- Simion T (2018) Breeding Cowpea Vignaunguiculata l. Walp for Quality Traits. Ann Rev Resear 3: 555609. Link: https://bit.ly/3rdTmql
- Ravelombola WS, Shi AN, Weng YJ, Motes D, Chen PY, et al.(2016) Evaluation of total seed protein content in eleven Arkansas cowpea [ Vigna unguiculata (L.) Walp.] lines. American Journal of Plant Sciences 7: 2288-2296. Link: https://bit.ly/2NEKuMR
- MoANR (Ministry of Agriculture and Natural Resource) (2016) Plant and Animal Health Regulatory Directorate. Crop variety register issue No. 20. Addis Ababa, Ethiopia. Link: https://bit.ly/3tJJDtC
- Tesfaye S, Tesfaye B, Keneni G, Amsalu B (2020) Genetic diversity for immature pod traits in Ethiopian cowpea [ Vigna unguiculata (L.) Walp] landrace collections. African Journal of Biotechnology 19: 171-182. Link: https://bit.ly/2Pjkjvu
- Vavilov NI (1951) The origin, variation, immunity and breeding of cultivated plants. Chronica. Botanica 13: 1-36.
- Collaborative Crop Research Program (CCRP) (2015) Collaborative Crop Research program. Cowpea stallholder workshop, Accra, Ghana.
- Belay F, Gebresilasie A, Meresa H (2017) Agronomic Performance Evaluation of Cowpea [ Vigna unguiculata (L.) Walp] Varieties in Abergelle District, Northern Ethiopia. Journal of Plant Breeding and Crop Science 9: 139-143. Link: https://bit.ly/3sbY7Ca
- Omoigui LO, Ishiyaku MF, Kamara AY, Alabi SO, Mohammed SG (2006) Genetic variability and heritability studies of some reproductive traits in cowpea [ Vigna unguiculata (L.) Walp.]. Afr J Biotechnol 5: 1191-1195. Link: https://bit.ly/3vN1BgB
- IBPGR (1983) International Board for Plant Genetic Resources. Descriptors for Cowpea. Rome, Italy. Link: https://bit.ly/312MxNZ
- SAS Institute Inc (2010) Statistical Analysis System, Version 9.2 SAS institute Inc.
- Burton GW, DeVane EM (1953) Estimating heritability in tall fescue (Festuca arundinaceae) from replicated clonal material. Agron J 45: 478-481. Link: https://bit.ly/3c9blKA
- Singh RK, Chaundry BD (1985) Biometrical Methods in Quantitative Genetic Analysis. 2nd ed., Kalayani Publishers, New Delhi-Ludhiana. Link:
- Allard RW (1960) Principles of plant breeding. New York; John Wiley and Sons Inc. 485. Link: https://bit.ly/2NENkBv
- Johnson HW, Robinson HF, Comstock RE (1955) Estimation of genetic and environmental variability in soybeans. Agron J 47: 314–318. Link: https://bit.ly/3vNj6O2
- Ward JR (1963) Hierarchical grouping to optimize and function. Journal of American Statistical Association 58: 236-244. Link: https://bit.ly/3tGFvdY
- Mahalanobis PC (1936) On the generalized distance in statistics. Proceeding of National Academic of Science 2: 49-55. Link: https://bit.ly/3cSVBKR
- Milligan GW, Cooper MC (1985) An examination of procedures for determining the number of cluster in data set. Psychometrika 50: 159-179. Link: https://bit.ly/3cYVOfg
- Sharama JR (1998) Statistical and Biometrical Techniques in Plant Breeding. New Delhi, India.
- Deshmukh SN, Basu MS, Reddy PS (1986) Genetic variability, character association and path coefficient analysis of Quantitative traits in Viginia bunch varieties of ground nut. Indian Journal of Agricultural Science 56: 515-518. Link: http://bit.ly/3cTYg6V
- Krishnan R, Lovely B, Suja G (2019) Genetic Variability and Heritability Studies in Cowpea (Vigna Unguiculata (L.) Walp). International Journal of Agricultural Science and Research 9: 311–316. Link: https://bit.ly/3tOYi77
- Manggoel W, Uguru MI, Ndam ON, Dasbak MA (2012) Genetic variability, correlation and path coefficient analysis of some yield components of ten cowpea [ Vigna unguiculata (L.) Walp] accessions. Journal of Plant Breeding and Crop Science 4: 80-86. Link: https://bit.ly/3lEKV6v
- Mofokeng MA, Mashilo J, Rantso P, Shimelis H (2020) Genetic variation and genetic advance in cowpea based on yield and yield-related traits. Acta Agriculturae Scandinavica, Section B- Soil & Plant Science 70: 381-391. Link: https://bit.ly/318ebZM
- Singh BD (2001) Plant Breeding: Principles and Methods 6th ed. Kalyani Publishers, New Delhi, India.
- Thorat A, Gadewar RD (2013) Variability and correlation studies in cowpea ( Vigna unguiculata ). International Journal for Environmental Rehabilitation and Conservation 4: 44-49. Link: https://bit.ly/3f2o837
- Khan H, Viswanatha KP, Sowmya HC (2015) Study of genetic variability parameters in cowpea ( Vigna unguiculata L. walp.) germplasm lines. Bioscan 10: 747–750. Link: http://bit.ly/315WWIQ
- Khanpara SV, Jivani LL, Vachhani JH, Kachhadia VH (2016) Genetic variability, heritability and genetic advance studies in vegetable cowpea [ Vigna unguiculata (L.) Walp.]. Electr J Plant Breed 7: 408–411. Link: https://bit.ly/3daGjRV
- Hermes AT, Carlos CA, Miguel EC, Darlin AP, Andrea GB (2018) Estimation of genetic parameters in white seed cowpea ( Vigna unguiculata (L) Walp). Austr J of Crop Sci 12: 1016-1022. Link: https://bit.ly/3tW8KK7
- Srinivas J, Kale VS, Nagre PK (2017) Evaluation of different cowpea varieties and genotypes. Int J Pure Appl Biosci 5: 329-334. Link: https://bit.ly/3rfNvkI
- Ubi EB, Mignouna H, Obigbesan G (2001) Segregation for seed weight, pod length and days to flowering following cowpea cross. Afr Crop Sci J 9: 463-470. Link: http://bit.ly/3tDjnB2
- Sharma M, Sharma PP, Upadhyay B, Bairwa HL, Meghawal DR (2017) Character association and path analysis in cowpea [ Vigna unguiculata (L.) Walp] Germplasm Line. Int J Curr Microbiol App Sci 6: 786–795. Link: https://bit.ly/2NHz6Qs
- Das RR, Talukdar P, Kumar P, Neog SB (2018) Relationship among different secondary traits and seed yield in cowpea ( Vigna unguiculata L. Walp). Internationl Journal of Current Microbiology and Applied Sciences 7: 1382–1396. Link: https://bit.ly/3vNZ6e9
- Walle T, Mekbib F, Amsalu B, Gedil M (2019) Genetic Diversity of Ethiopian Cowpea [ Vigna unguiculata (L) Walp] Genotypes Using Multivariate Analyses. Ethiopian J Agric Sci 29: 89-104. Link: https://bit.ly/3c879dW
- Molosiwa OO, Chiyapo G, Joshuah M, Stephen MC (2016) Phenotypic variation in cowpea ( Vigna unguiculata [L.] Walp.) germplasm collection from Botswana. International Journal of Biodiversity and Conservation 8: 153-163. Link: https://bit.ly/3vWz5cP
- Triveni MS, Anjali M, Neeraj T (2018) Morphological based genetic diversity studies of cowpea. Plant Archives 18: 227-231.
- Diouf D, Hilu KW (2005) Microsatellites and RAPD markers to study genetic relationships among cowpea breeding lines and local varieties in Senegal. Genetic Resources and Crop Evolution 52: 1057-1067. Link: https://bit.ly/3f26qNe
- Dwevedi KK, Lal GM (2009) Assessment of genetic diversity of cultivated chickpea (Cicer arietinumL.). Asian Journal of Agricultural Science 1: 7-8.
- Jivani DR, MS Mehta, Pithia RB, Madariya C, Mandavia K (2013) Variability analysis and multivariate analysis in chickpea (Cicer arietinum L.). Electronic Journal of Plant Breeding 4: 1284-1291. Link: http://bit.ly/3cSXeYZ
- Ragab DD, Babiker EE, Eltinay AH (2004) Fractionation, solubility and functional properties of cowpea ( Vigna unguiculata ) proteins as affected by pH and/or salt concentration. Journal of Food Chemistry 84: 207-212. Link: http://bit.ly/31iJQYR
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
