International Journal of Agricultural Science and Food Technology
Evaluation of elite rice lines for resistance to Kenya blast fungus (Magnapothe Oryzae)
Roselyne U Juma1*, Rosemary Murori2, Felister N Mbute3, Douglas W Miano3 and Joshua N Cobb4
2International Rice Research Institute, (IRRI) Kenya, ILRI C/O IRRI PO Box 30709, Nairobi 00100, Kenya, Old Naivasha Road, Kenya
3University of Nairobi, (UON) Upper Kabete Campus 29053, – 00625, Kangemi, Kenya
4RiceTec. Inc, PO Box 1305, Alvin, TX 77512, Kenya, USA
Cite this as
Juma RU, Murori R, Mbute FN, Miano DW, Cobb JN (2024) Evaluation of elite rice lines for resistance to Kenya blast fungus (Magnapothe Oryzae). Int J Agric Sc Food Technol 10(2): 053-060. DOI: 10.17352/2455-815X.000207Copyright License
© 2024 Juma RU, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Blast resistance tends to often break down, these necessitate search-resistant genes. The screen house experiment was conducted in 2019 and 2020. A total of 56 rice genotypes, (Elite lines, monogenic lines, and local), were screened against ten Kenya isolates of Mangnaporthe oryzae. The establishment was by direct seeding of previously sprouted seeds. A completely Randomized Design (CRD) with two replicates was adopted. Inoculation was done 21 days after planting. Plants were maintained in the moist chamber (26–28 0C) for 48 hrs, then moved into an incubation chamber (25 ◦C ± 2). Disease assessment commenced 48 hours after inoculation until 21 days to full infection. Disease evaluations were performed according to the Standard Evaluation System of IRRI. Polymerase Chain Reactions (PCR) were carried out, blast resistant loci were identified using varied genetic markers that co-segregate with specific resistant loci
Analysis was implemented in R version 3.3.2 (R Core Team, 2016). BLUPs were computed, in a mixed model to estimate the random effect of the genotypes. The data was subjected to analysis of variance and means separated by Tukey’s test at P < 0.05. Cluster analyses were performed in JMP software version 11.2 for Windows. Seven genotypes (IR12A311, IR10M210, IR74, 1R02A127, IR09A130, R66 and IR10N230) were more susceptible, IR13N152 and IR14F711 were more resistant to all Isolates. The monogenic lines IRBLsh-B and IRBLkh-K3 which carry blast-resistant genes, were susceptible to at least seven Kenya isolates. The local checks (Basmati 370, BAS 270 (pi9), BW196, and BAS370 (pi9) were highly susceptible. The genotype IR12F711 was consistently resistant to all Kenya Isolates.
Introduction
Rice (Oryza sativa L.) is one of the important cereals contributing significantly to the world’s food sufficiency, this is true because more than half the world’s population consumes rice as the main staple [1]. Its productivity is usually affected by many major and minor diseases [2], Among them, the blast caused by Magnaporthe oryzae (synonym-Pyricularia oryzae) [3] is the most severe resulting into great losses globally.
The fungus is a major threat to rice production in all rice-growing ecologies in Kenya, causing enormous losses [4]. The disease causes yield losses of about 60% - 100% when an outbreak occurs [5]. In favorable conditions, this disease can devastate entire rice plants within 15 to 20 days.
Rice blast can destroy enough rice to feed more than 60 million people worldwide. The disease infects all the developmental stages of the plant producing symptoms on the leaf, collar, neck, panicle, and even in the glumes [6].
Since rice blast disease was identified in Kenya, several rice blast outbreaks have been experienced in rice irrigation schemes, causing more than 50% yield losses in some seasons. This is associated with intensive and continuous growth of susceptible popular varieties which enhances opportunities for the evolution of new pathotypes.
Farmers in the Mwea irrigation scheme have ever experienced rice blast disease destruction of 5600 hectares [5] in one season, this is equivalent to 10% to 20% of annual output. The average yield per hectare in the Mwea irrigation scheme is usually 6.5 tons of paddy rice, but due to frequent blast disease outbreaks, this may be reduced to 3 tons. Severe blast infections have been reported on Basmati 217, Basmati 370, and ITA 310 rice varieties across the rice-growing regions in Kenya [7].
There are variations of blast pathogens prevalent in Kenya which should be considered when testing for strain virulence. The study by Fukuta, et al. [8] targeting 23 resistance genes, reported that blast isolates from the four rice geographical regions (Western, Nyanza, Central, and Coast) in Kenya are different. This study identified three different classes. The classification was about the reaction of the rice blast isolates to the susceptible variety Lijiangxintuanheigu (LTH) and also to the differential rice lines that carry blast-resistant genes. The prevalence of rice blast races is influenced by many factors including variation in virulence due to genetic drift and gene flow of the blast fungus, environmental conditions, and cropping system, among others [9].
Control methods practiced include preventive measures, fertilizer management, and chemical application. These methods are costly and environmentally hazardous. The most effective and eco-friendly method for rice blast control is the use of rice cultivars with complete resistance [10]. However, the resistance in the cultivars breaks down after a few years of their release due to the abundance of new races of rice blast fungus virulent to the resistance. The rapid genetic evolution of the fungus often overcomes the resistance conferred by major genes after a few years of intensive agricultural use [11]. Developing similar resistant genes widely in rice varieties results in uniformity, which finally causes the corresponding M. oryzae strain to undergo mutations. Such mutations lead to the occurrence of new pathogenicity, often resulting in disease outbreaks overcoming the specific blast resistance genes [12]. Further, blast-resistant genes express varied trends of resistance to different M. oryzae [13]. Knowledge of population genetics of rice genotypes is important for the selection of parental lines used as resistant sources.
To effectively utilize resistance, mixed host resistance with different complete resistance genes to blast is highly recommended. This multiple lines approach has been demonstrated to be effective [14] as compared to line resistant variety approach [15,16]. Although race-specific resistance is powerful and effective against particular pathotypes, it often breaks down. Due to this reason, other than utilizing broad-spectrum R genes, it is vital to identify the distribution frequency of these known R genes in varieties in a particular area and then design varieties by pyramiding appropriate R genes. The application of combinations of different genes and pyramiding them in a single rice cultivar for developing broad-spectrum durable resistance is the most desired strategy [11]. Therefore, it is important to continuously enrich the resistance reservoir in the country to effectively manage the blast disease [17].
The International Rice Research Institute (IRRI) has developed rice varieties with resistance to blast disease, and through collaborative research, the Kenya rice research program has obtained some of the lines for screening against the blast strains in Kenya. We hypothesize that blast-resistant genes in the global elite varieties will be effective in controlling the disease.
Objective
The present study was undertaken to determine the resistance of the global elite lines to the Kenyan M. oryzae and select parental breeding lines
Materials and methods
Materials and methods
The study was conducted in the blast screen house nursery at the International Livestock Research Institute (ILRI) research facility in Nairobi, Kenya in 2019 and 2020.
Test material
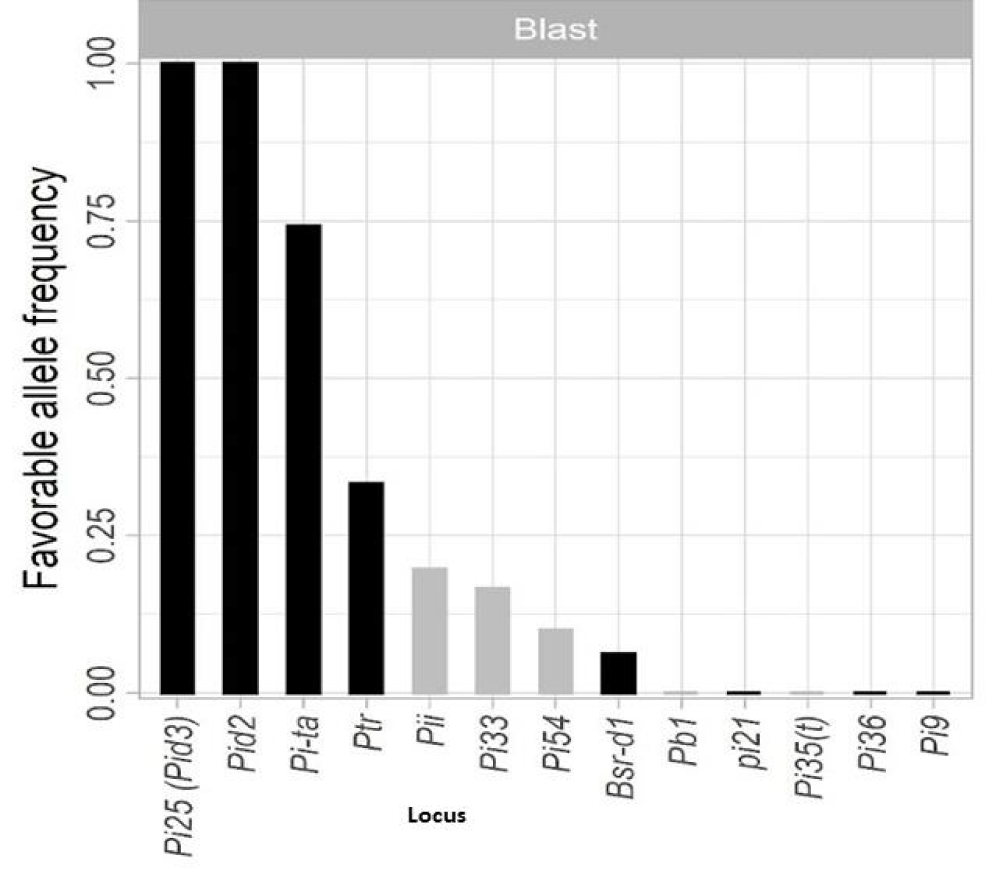
The panel of 61 test genotypes (Table.S1), consisted of, 51 Elite parental genotypes obtained from the Irrigated Breeding Program of the International Rice Research Institute (IRRI). Selections were made based on adaptability to the Kenyan irrigated ecology. The elite genotypes are known to have high allele frequency (Figure 1); (> 95%) for (Pita, Pi25 (Pid3), and Pid2; moderate frequency (< 70%) (Pita); low frequency (< 50% Bsr-d1, Pi33, Pi54, Pii, and Ptr) but absent in (Pi9, Pi35, Pi21) alleles [18], additional 5 monogenic genotypes were included as a control for isolate specific rice blast resistance (Table S2), the cultivar Lijiangxintuanheigu (LTH) was also included as a susceptible check. Further, five local checks from Kenya; BAS 370, BW 196, BAS217 (Pi9), BAS370 (Pi9), and ITA 310 were added.
Blast isolates
The ten isolates (KE0001, KE0002, KE0016, KE0018, KE0215, KE0218, KE0381, KE0401, KE0442, KE0495) of M. oryzae fungi used in this study is a subset of Kenyan isolates within the larger collection (n = 250; enrichment of the biobank ongoing) which were made by Durable Rice Blast Resistance for sub-Saharan Africa through a support of Biotechnology and Biological Society Research Council (BBSRC, UK). Collections are stored in a repository in BecA-ILRI. The subset was selected based on virulence and regional diversity as previously reported [19].
Design and model
A Completely Randomized Design (CRD) was adopted, with two replicates.
Statistical model: Yij =µ + Ti + еij where,
- Yij – is the observation of genotype I in j replication
- μ overall mean based on all observations
- Ti effect of genotypes
- еij Error term assumed to be independent and normally distributed with mean zero and constant variance for each
Hypothesis:
Ho: T1= T2 = …= Tv
Ha: All Tis are not equal.
Conidial suspension preparation
Preparation was according to [20,21]. In summary, the previously isolated fungus that was trapped on a filter paper and stored under dry freeze was revived on Malt Extract Agar (MEA) under a Biosafety Cabinet chamber. The petri plates with the isolate were incubated at 25 °C for at least 3days. The fungus was transferred to the Rice Bran Agar (RBA) and prepared in advance for induction of sporulation. The RBA plates were incubated under 12 light-hour conditions and 12-hour darkness at 25 °C to induce sporulation for 18 days. Clean RBA petri plates were used to prepare the inoculum. Ten milliliters of double distilled water was poured into each petri plate with confirmed sporulation, and suspended by scrubbing off the mycelia surface using a soft brush while concentrating the solution containing conidia on each incubated plate of the same isolate of all the petri plates. The solution was homogenized in a beaker and filtered using cheesecloth to trap the mycelia in a sterile beaker of an appropriate volume. Using a hemocytometer the concentration of spores in the inoculum was estimated by counting the spores using a cell counter to a final concentration of 4 × 105 conidia/ml in distilled water. To enhance the adherence of conidia to rice leaves, 20ml Tween 20 in 0.2% gelatin was added to the suspension for improved adhesion of the blast spores on the inoculated plant.
Establishment and inoculation of test plants
The process for inoculation was as in Figure 2, where the lines for inoculation were grown in 15 cm × 20 cm × 5cm cylindrical pots filled with potting media (forest soil, river sand, and manure at a 2:1:1 ratio) in a Completely Randomized Design (CRD) replicated twice. Test lines were established by direct seeding of 12 previously sprouted seeds. Diamonium Phosphate (DAP) fertilizer was applied to the newly emerged seedlings at 10 gms per pot. Plants were inoculated at 21 days after planting. After inoculation, the plants were maintained in a moist chamber at 26–28 °C for 48 hrs, after which the plants were transferred to the incubation chamber at 25 °C ± 2 for seven days before disease assessment. Ten plants were randomly selected for evaluation.
Disease assessment
Disease evaluations for each test line were performed according to the Standard Evaluation System of IRRI [22], for screening leaf blasts, with scores ranging from 0 to 9 (Figure 3). Ten plants were observed and diseased plants were identified, blast lesion type and the leaf area covered with infection were recorded.
Disease incidence and severity
Disease incidence was estimated as the proportion of infected plants and assigned a numeric score. The average of the numerical scores was considered the blast disease score for the particular genotype
I = ∑x/N
Where X is the sum of numeric scores for the number of diseased plants over the total number (N) of plants evaluated, the assessment for the disease was recorded at three intervals from the time of inoculation (0, 7, and 21) days after inoculation (DAI) for each genotype.
- Disease symptom incidence was observed for 10 randomly picked plants. The disease incidence was measured using 0 – 9 standard scoring.
The disease severity on the other hand was by accurately assessing the lesion area of the leaf or the proportion of diseased area to the entire plant. The severity data were used to determine the area under the disease progress curve (AUDPC) for each genotype.
Calculating Area under Disease Progression Curve (AUDPC) for Elite rice Germplasm
The AUDPC was estimated using the midpoint method as expressed below:
Where;
xi = the proportion of host tissue damaged at the ith day,
ti = the time in days after the appearance of the disease at the ith day,
n = the total number of observations.
The AUDPC was calculated in such a way that the first and second scores are added, multiplied by the time between them, and divided by two, this is repeated for the second and third scores and then divided by 2. The procedure is repeated for all consecutive pairs of scores and the values are added together to determine the AUDPC.
Lower AUDPCs represent slower disease progression and greater resistance to the disease. Higher AUDPCs represent faster disease progression and higher susceptibility to the disease.
Molecular characterization of elite parents
The elite parental lines phenotyped for resistance to Kenya rice blast isolates were genotyped using gene-based markers to confirm or identify any blast-resistant allele.
Genomic DNA extraction
The elite parental lines were planted in a greenhouse and leaf samples were obtained 21 days after planting for genomic DNA isolation. Leaf samples were collected in 2 ml microcentrifuge tubes, 1.2 ml of 2 × CTAB buffer was added, and the samples were incubated at 65 °C for 30 min. After the addition of 800µl of phenol:chloroform: isoamyl alcohol (25:24:1) and mixing, the sample tubes were centrifuged for 15 min at 13,000 rpm. The aqueous supernatant was collected in new 1.5 ml tubes and was treated with RNaseA at 37 °C for 30 min. DNA was precipitated by adding 560µl of isopropanol and pelleted by centrifugation at 13,000 rpm for 10 min. After washing twice with ethanol, the dried DNA pellet was dissolved in 50µl of 1 × TE buffer and was used for Polymerase Chain Reaction (PCR) analysis.
PCR amplification
Polymerase chain reaction (PCR) was carried out to determine the presence/absence of blast-resistant loci using varied genetic markers that co-segregate with specific resistant loci. Forward and reverse primers were used to amplify the targeted fragment of ∼2 kb from the test genotypes (Table S3). The PCR program included initial denaturation at 95 °C for 5 min; followed by additional denaturation at 95 °C for 30 s, annealing at 51 °C for 30 s, and extension at 72 °C for 1 min (these three steps were repeated for 39 cycles); followed by final extension at 72 °C for 7 min. DNA was precipitated by adding 560µl of isopropanol and pelleted by centrifugation at 13,000 rpm for 10 min. After washing twice with ethanol, the dried DNA pellet was dissolved in 50µl of 1 × TE buffer and was used for PCR analysis. The PCR products were electrophoresed in 3% agarose gels at 100 V for 3-4 h. The gel was scanned and photographed with the DNR MiniBis Bioimaging System, the obtained banns were scored in 1,0 binary matrix, scored as present (1) or absent (0), for further analysis [23].
Statistical analysis
The statistical analysis was implemented in R version 3.3.2 (R Core Team, 2016). The computation of Blups was estimated in a mixed model to estimate the random effect of the genotypes. The data was subjected to analysis of variance to determine whether or not there is a statistically significant difference between the genotypes in blast incidence, significant differences between means were identified by Tukey’s test at p < 0.05.
Cluster analyses were performed by Ward’s hierarchical method [24] with the software JMP version 11.2 for Windows (2014; SAS Institute, Inc., Cary, NC, USA), based on the gel data of the SNP, SSR, InDel, LM, and FM markers.
Using the allele sizes obtained with each marker for the 61 rice genotypes for the input matrix for genetic analysis in binary format. The 0/1 matrix was used to calculate the similarity between the elite lines. The resultant similarity matrix was employed to construct dendrograms using Agglomerative Hierarchical clustering. An ANOVA was conducted using the cluster groups as predictors to find the significant difference of the clusters on blast disease reaction.
Results
Overall genotypic means for the response of elite lines to Kenya blast isolates
A one-way ANOVA was performed to compare the effect of Kenyan rice blast isolates on the Elite rice genotypes. The one-way ANOVA revealed that there was a statistically significant difference in rice elite genotypes at p < 001, the observed p-value from the ANOVA table is less than 0.05, indicating that there is enough evidence to conclude that the genotype means are not equal. Similarly, the means for blast isolates reaction was highly significant and the interaction of genotype and the blast isolates was also highly significant (Table 1).
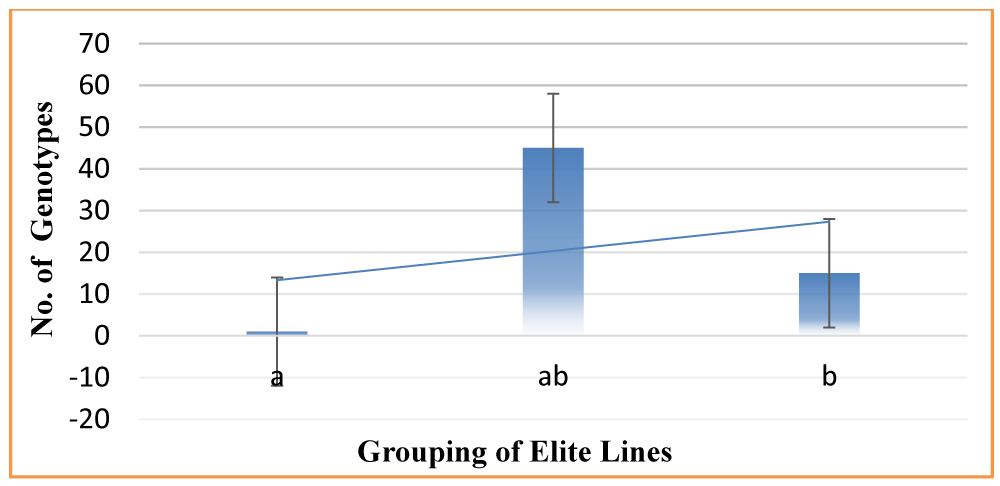
Tukey’s HSD Test for multiple comparisons found that the mean values of rice elite genotypes were significantly different between Basmati370, IR4A115, and IR12A311 at 95% probability. The entire panel was grouped into three main categories (Figure 4).
Reaction of individual blast Isolate with the whole test panel
The reaction for the global genotypes followed a similar trend for most Isolates, where very few lines were completely susceptible, similarly for complete resistance, and moderate reaction. Seven of the genotypes (IR12A311, IR10M210, IR74, and 1R02A127, IR09A130, R66, and IR10N230) were more susceptible, and lines IR13N152 and IR14F711 were more resistant to all isolates. Although the monogenic lines IRBLsh-B and IRBLkh-K3 carry some blast-resistant genes, they however showed susceptibility to at least seven Kenya isolates. The local checks (Basmati 370, BAS 217 (pi9), BW196, and BAS370 (pi9) were highly susceptible. Similarly, in an earlier study [18] this panel was evaluated against five Phillipines blast isolates of Magnaporthe oryzae under controlled conditions. Based on the phenotypic measurement, the most virulent isolate was M64-1-3-9-1 and the least virulent was CA89. A wide variation in the resistance to the five blast isolates was found in the panel lines with most of the genotypes displaying intermediate resistance to one or more isolates. Surprisingly, the most resistant genotypes to one isolate were found not to be the most resistant to the other isolates, suggesting host-isolate interaction. Interestingly genotype IR14F711 which showed resistance consistently to all Kenya isolates was also resistant to the Philippines’ isolates (CA89 and 1K81-25) [18]. Based on the gel analysis it was revealed that this genotype carries at least more than ten blast-resistant alleles.
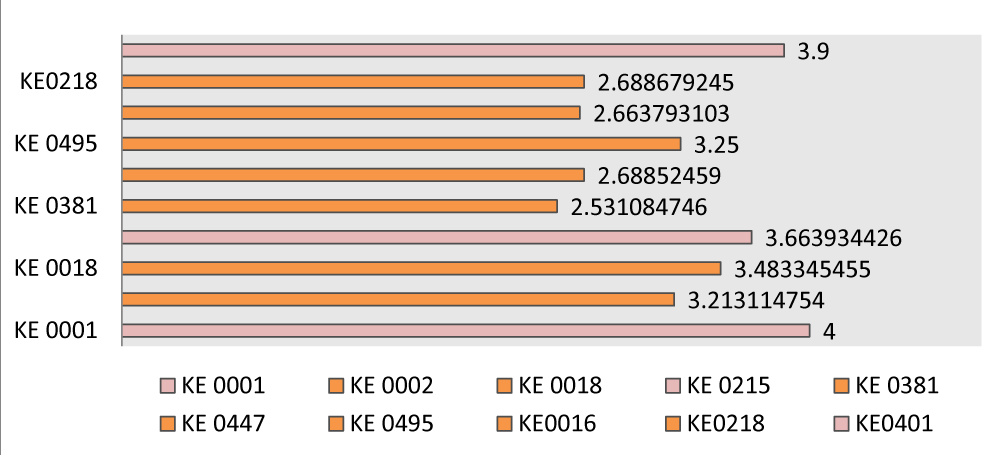
Most of the genotypes were susceptible to three of the isolates (KE0001, KE0215 & KE0401), followed by the next three (KE0002, KE0018 & KE0447) of which the genotypes were moderately susceptible (Figure 5).
Tested genotypes clustered into 4 groups (Resistant, Moderately Resistant, Susceptible, and Highly Susceptible).
Disease progress
Conducive conditions facilitated early disease initiation, rapid disease development, and the highest disease pressure in the susceptible genotypes during testing. Delayed disease initiation, slower rates of disease development, and lower final disease severities were recorded for some of the elite lines. Disease severities in the susceptible genotypes were significantly higher than those in the less susceptible genotypes.
Grouping of the Elite panel based on gel
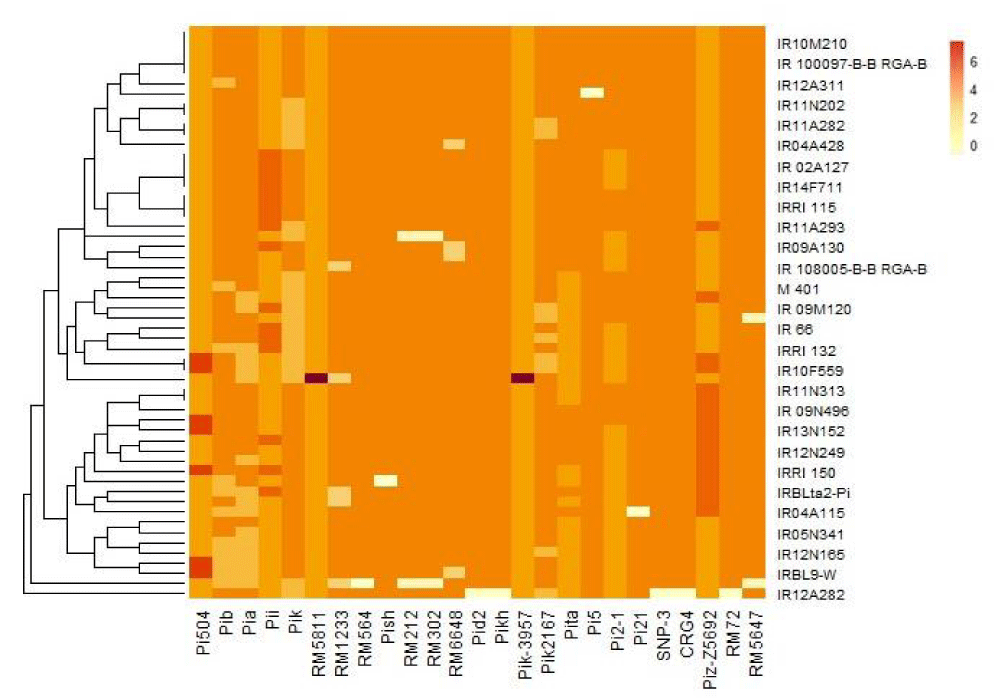
Cluster analysis based on gel genotyping of blast-resistant genes in the elite panel lines revealed three main clusters, which further branched into six sub-clusters (Figure 6). These clusters varied in size (3 to 14 lines per cluster) and all clusters were not different in terms of blast-resistant genes. The cluster groups were found not to be significantly difference between clusters (p -value > 0.05).
It is most likely that the phenotypic variation in the elite lines is due to the favorable allele combinations for genes associated with blast resistance. Understanding the genetic makeup of the elite is important especially when being targeted as parental lines. The grouping of lines was according to the clustering as per blast-resistant alleles (Table 2). Alleles pid2, Pish, and Pikh are fixed in the elite lines, and the combination of Pi-ta, Pikh, Pi2, Pish, Pid, Pia, and Pi21 was associated with resistant lines. Five genotypes (R13N157, IR 09N496, IR13N102, IR10M210, and IRRI 115) were most resistant with at least 20 resistant Alleles including Pita.
Discussion
A total of 61 rice genotypes from the global breeding program (IRRI) consisting of Elite, Trait donors, and monogenic lines and some local checks from Kenya, showed varied resistance.
The Basmati 217, Basmati 370, and ITA 310 were very susceptible to most isolates, this finding agrees with Kariaga, et al. [5] and Kihoro, et al. [5,7]. The accessions of Basmati 217 and Basimti 370 inbuilt by Pi9 were susceptible to the strains of blast. It is most likely that the allele gene could be less effective because, from the gel analysis, the accessions have this gene.
Nevertheless, serious infections have already been reported in Basmati 217, Basmati 370, and ITA 310, and have become a big problem for rice cultivation in Kenya [5,7]. Basmati 217 and Basmati 370 harbored Pi20(t) and one of Pik allele genes or Pi3. This research is limited only to evaluation under a controlled environment and only leaf blast was accessed
Recommendation and Conclusion
The elite genotypes carry the desired variability for blast resistance, at least 40% of the lines were resistant to either of the Kenya blast isolates. However, single gene isolates may not confer complete resistance to all Kenya blast isolates. The resistant alleles in these lines are quite effective in protecting the lines against the Kenya blast fungus. Therefore, the rice elite lines can make good parental lines to improve blast resistance in the Kenya rice population.
The authors are grateful to IRRI for providing the study material and space for screen housework. We are thankful to ILRI, especially Dr. Samuel Mutiga, for providing the Kenyan blast isolates and screen house space for phenotyping the material. The Bill and Melinda Gates Foundation is most appreciated for funding this work.
- Fairhurst T, Dobermann A. Rice in the Global Food Supply. Better Crops Int., 2002; 16.
- Dianga AI, Musila RN, Joseph KW. Rainfed Rice Farming Production Constrains and Prospects, the Kenyan Situation. IntechOpen. 2021. https://doi.org/10.5772/intechopen.98389
- Zhang N, Luo J, Rossman AY, Aoki T, Chuma I, Crous PW, Dean R, de Vries RP, Donofrio N, Hyde KD, Lebrun MH, Talbot NJ, Tharreau D, Tosa Y, Valent B, Wang Z, Xu JR. Generic names in Magnaporthales. IMA Fungus. 2016 Jun;7(1):155-9. doi: 10.5598/imafungus.2016.07.01.09. Epub 2016 Jun 8. PMID: 27433445; PMCID: PMC4941683.
- Onyango AO. Rice production, Food security, New rice for Africa, Agricultural research and development. World Environment. 2014; 4(4):172–179.
- Kihoro J, Bosco NJ, Murage H, Ateka E, Makihara D. Investigating the impact of rice blast disease on the livelihood of the local farmers in greater Mwea region of Kenya. Springerplus. 2013 Jul 10;2(1):308. doi: 10.1186/2193-1801-2-308. PMID: 23888278; PMCID: PMC3715686.
- Simkhada K, Thapa R. Rice Blast, A Major Threat to the Rice Production and its Various Management Techniques. Turkish Journal of Agriculture - Food Science and Technology. 2022; 10:147–157. https://doi.org/10.24925/turjaf.v10i2.147-157.4548
- Kariaga M. Identification of Rice Blast (Pyricularia oryzae Cav.) Races from Kenyan Rice Growing Regions Using Culture and Classical Characterization. Journal of Research in Agriculture and Animal Science. 2016; 2321–9459.
- Fukuta Y, Telebanco-Yanoria MJ, Hayashi N, Yanagihara S, Machungo CW, Makihara D. Pathogenicities of Rice Blast (Pyricularia oryzae Cavara) Isolates From Kenya. Plant Disease. 2019; 103(12): Article 12. https://doi.org/10.1094/PDIS-04-19-0870-RE
- Zhan J, Thrall PH, Papaïx J, Xie L, Burdon JJ. Playing on a pathogen's weakness: using evolution to guide sustainable plant disease control strategies. Annu Rev Phytopathol. 2015;53:19-43. doi: 10.1146/annurev-phyto-080614-120040. Epub 2015 May 4. PMID: 25938275.
- Ashkani S, Rafii MY, Shabanimofrad M, Miah G, Sahebi M, Azizi P, Tanweer FA, Akhtar MS, Nasehi A. Molecular Breeding Strategy and Challenges Towards Improvement of Blast Disease Resistance in Rice Crop. Front Plant Sci. 2015 Nov 16;6:886. doi: 10.3389/fpls.2015.00886. PMID: 26635817; PMCID: PMC4644793.
- Younas MU, Wang G, Du H, Zhang Y, Ahmad I, Rajput N, Li M, Feng Z, Hu K, Khan NU, Xie W, Qasim M, Chen Z, Zuo S. Approaches to Reduce Rice Blast Disease Using Knowledge from Host Resistance and Pathogen Pathogenicity. Int J Mol Sci. 2023 Mar 5;24(5):4985. doi: 10.3390/ijms24054985. PMID: 36902415; PMCID: PMC10003181.
- Wu W, Wang L, Zhang S, Li Z, Zhang Y, Lin F, Pan Q. Stepwise arms race between AvrPik and Pik alleles in the rice blast pathosystem. Mol Plant Microbe Interact. 2014 Aug;27(8):759-69. doi: 10.1094/MPMI-02-14-0046-R. PMID: 24742074.
- Bagnaresi P, Biselli C, Orrù L, Urso S, Crispino L, Abbruscato P, Piffanelli P, Lupotto E, Cattivelli L, Valè G. Comparative transcriptome profiling of the early response to Magnaporthe oryzae in durable resistant vs susceptible rice (Oryza sativa L.) genotypes. PLoS One. 2012;7(12):e51609. doi: 10.1371/journal.pone.0051609. Epub 2012 Dec 12. PMID: 23251593; PMCID: PMC3520944.
- Zhu Y, Chen H, Fan J, Wang Y, Li Y, Chen J, Fan J, Yang S, Hu L, Leung H, Mew TW, Teng PS, Wang Z, Mundt CC. Genetic diversity and disease control in rice. Nature. 2000; 406(6797):6797. https://doi.org/10.1038/35021046
- Keesing F, Holt RD, Ostfeld RS. Effects of species diversity on disease risk. Ecol Lett. 2006 Apr;9(4):485-98. doi: 10.1111/j.1461-0248.2006.00885.x. PMID: 16623733.
- Raboin LM, Ramanantsoanirina A, Dusserre J, Razasolofonanahary F. Two‐component cultivar mixtures reduce rice blast epidemics in an upland agrosystem. 2012; 6:1103–1111. https://doi.org/10.1111/j.1365-3059.2012.02602
- Thon MR, Pan H, Diener S, Papalas J, Taro A, Mitchell TK, Dean RA. The role of transposable element clusters in genome evolution and loss of synteny in the rice blast fungus Magnaporthe oryzae. Genome Biol. 2006;7(2):R16. doi: 10.1186/gb-2006-7-2-r16. Epub 2006 Feb 28. PMID: 16507177; PMCID: PMC1431731.
- Juma RU, Bartholomé J, Thathapalli Prakash P, Hussain W, Platten JD, Lopena V, Verdeprado H, Murori R, Ndayiragije A, Katiyar SK, Islam MR, Biswas PS, Rutkoski JE, Arbelaez JD, Mbute FN, Miano DW, Cobb JN. Identification of an Elite Core Panel as a Key Breeding Resource to Accelerate the Rate of Genetic Improvement for Irrigated Rice. Rice (New York, N.Y.). 2021; 14(1): 92. https://doi.org/10.1186/s12284-021-00533-5
- Mutiga SK, Rotich F, Ganeshan VD, Mwongera DT, Mgonja EM, Were VM, Harvey JW, Zhou B, Wasilwa L, Feng C, Ouédraogo I, Wang GL, Mitchell TK, Talbot NJ, Correll JC. Assessment of the Virulence Spectrum and Its Association with Genetic Diversity in Magnaporthe oryzae Populations from Sub-Saharan Africa. Phytopathology. 2017 Jul;107(7):852-863. doi: 10.1094/PHYTO-08-16-0319-R. Epub 2017 May 5. PMID: 28368237.
- Miura Y, Ding C, Ozaki R, Hirata M, Fujimori M, Takahashi W, Cai H, Mizuno K. Development of EST-derived CAPS and AFLP markers linked to a gene for resistance to ryegrass blast (Pyricularia sp.) in Italian ryegrass (Lolium multiflorum Lam.). Theor Appl Genet. 2005 Sep;111(5):811-8. doi: 10.1007/s00122-005-0001-8. Epub 2005 Oct 18. PMID: 16025307.
- Molinari C, Talbot NJ. A Basic Guide to the Growth and Manipulation of the Blast Fungus, Magnaporthe oryzae. Curr Protoc. 2022 Aug;2(8):e523. doi: 10.1002/cpz1.523. PMID: 35997707; PMCID: PMC9542439.
- IRRI I. Standard Evaluation System (SES). http://www.knowledgebank.irri.org/images/docs/rice-standard-evaluation-system.pdf 2002.
- Singh A, Kumar R, Singh A, Bansal S, Singh D, Tomar A. Studies on Genetic Variability in Rice Using Molecular Markers. Vegetos-An International Journal of Plant Research. 2011; 24(1): Article 1.
- Ward JH, Hook ME. Application of a Hierarchical Grouping Procedure to a Problem of Grouping Profiles. Educational and Psychological Measurement. 1963; 23(1): 69–81. https://doi.org/10.1177/001316446302300107
Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.




 Save to Mendeley
Save to Mendeley
