International Journal of Aquaculture and Fishery Sciences
Prevalence and Antimicrobial Susceptibility of Pathogenic Bacteria in Nile Tilapia, Oreochromis niloticus L
Begonesh Bekele, Kassaye Balkew Workagegn* and P Natarajan
Cite this as
Bekele B, Workagegn KB, Natarajan P (2019) Prevalence and Antimicrobial Susceptibility of Pathogenic Bacteria in Nile Tilapia, Oreochromis niloticus L. Int J Aquac Fish Sci 5(4): 022-026. DOI: 10.17352/2455-8400.000047Nile tilapia, Oreochromis niloticus, is one of the most popular aquaculture fish species in the world. However, among several challenges, the presence of pathogenic bacteria causes high economic losses. Thus, the main objective of this study was to isolate and identify the potent bacterial pathogens from Nile tilapia reared at Hawassa Fish Research and Multiplication Station, Ethiopia. For this, infected fish samples were collected from the research station and subjected to microbiological and biochemical tests. The results of the study revealed that 75% of fish with the length group ranged from 14-17.9cm; 52% with the length group ranged from 18-21.9 cm and 33% with the length group ranged from 22-26.9cm were infected by different bacteria belonging to the genera Vibrio, Escherichia, Aeromonas, Pseudomonas, Salmonella and Streptococcus. Except for Streptococcus, all isolates belonged to gram negative bacteria. The bacterial population observed in fish organs was significantly high in intestine (12.43±0.55 Log10 CFU-1g) than in liver (6.48±1.06 Log10 CFU-1g). An antibiogram test showed that isolated bacteria were sensitive to gentamycin, tetracycline and amoxicillin. In conclusion, the present results clearly indicate that cultivable fishes are prone to infection by infectious and non-infectious and that it may affect fish and their product quality which leads to economic loss and livelihood of farmers who depend on small scale aquaculture.
Introduction
The world’s population is growing rapidly, being expected to reach 9.8 billion by 2050 [1]. To feed this population, food production must increase by 60% worldwide [2]. However, food production from the agricultural sector will fail to meet such high demand due to climatic effects. Thus, climate-smart aquaculture is vital to increase food production. It is one of the primary sources of cheap animal protein for the rapidly growing human population. According to FAO [3], aquaculture production has increased from 61.8 million tons in 2011 to 80 million tons in 2016. The bulk of this production came mainly from Asia and Latin America. Asia accounted for about 88.9% of the world aquaculture production. Although the contribution of African aquaculture to the world fish production is relatively low, it increased from 0.49% in 1995 to 2.3% in 2014 [4]. Aquaculture in Ethiopia remains more potential than the actual practice, even though the country’s environmental conditions support its development. Extensive and semi-intensive aquaculture, in the form of stocking and enhancing artificial lakes, reservoirs and small water bodies, has been practiced since 1975 through the Sebeta National Fishery and Other Aquatic Life Research Center (SNFARC).
Although the country has rich fish biodiversity, the most dominant cultured fish species are Nile tilapia (Oreochromis niloticus), catfish (Clarias gariepinus) and common carp (Cyprinus carpio). Among these, Nile tilapia contributes about 50% of the overall fish production in the country [5,6]. This is because Nile tilapia has suitable cultivable characteristics such as efficient use of natural and artificial feeds, resistance to diseases, tolerance to a wide range of environmental conditions, relatively fast growth rate, and excellent meat quality. In semi-intensive and intensive aquaculture production systems, however, disease is a primary constraint that affects the growth of many cultivable fish species, and is responsible for hampering production and expansion of the sector and thereby reducing socioeconomic development of many developing countries of the word. For instance Asia has been faced with mass mortalities of many cultured fishes due to the occurrence of different bacterial diseases such as Aeromonas, Vibrio, and Pseudomonas [7]. In most cases, they cause inflammation, ulcer and hemorrhages that lead to reduce the quality of fish and fish products. Noninfectious diseases due to pollution, algal toxins feed contamination and water quality also common in cultured fishes which can have devastating effects on fish growth occasionally leading to crop loss [8]. Therefore, the main objective of this study was to isolate and identify the potential pathogenic bacterial in Nile tilapia, and to test their drugs sensitivity.
Materials and Methodology
Description of the study area
Fish samples were collected in 2018 from the Fish Breeding Station of the Hawassa Agricultural Research Center (HARC), 273 km south of Addis Ababa, Ethiopia. Geographically the site lies between 60 55’ 0’’ to 70 6’ 0’’latitude north and 38025’ 0’’ to 380 34’ 0’’ longtudes east and is situated 1,686m above mean sea level (Figure 1).
Preparation of fish samples for bacterial isolation
Sixty Nile tilapia were collected from the Fish Breeding Station of the HARC. Collected fish samples were held in plastic bags and taken to Hawassa University Veterinary Laboratory for the isolation and identification of pathogenic bacteria. Nile tilapia with body length ranged from 14cm and 21cm and body weight ranged from 43.2g and 139g were collected from the pond. For both external and internal examination, incision on the body surface was made by using a sterile scalpel. Immediately, the sample was placed in 70% ethanol. From these, bacteria were isolated from the body surface, gills, liver, intestine and kidney of fish aseptically.
Preparation of serial dilution and incubation
One gram samples from the above mentioned organs of each fish were taken and mixed with 100ml of 0.1% sterile peptone water in sterile bottles. Similarly, mixed with 9ml of 0.1% sterile peptone water in sterile test tubes. The bottles were then shaken thoroughly, and a 10-fold serial dilution was carried out. In this regard, 1ml of the original mixed sample was transferred to the first test tube and mixed thoroughly. From this solution, 1 ml was taken from the first test tube and added to the second test tube and mixed thoroughly. This procedure continued until the tenth serial dilution. Later, from each serially diluted sample, 0.1ml was transferred to nutrient agar using a pipette and dispensed and cultured by the glass spread method [9]. All the incubated nutrient agar plates were incubated in an inverted position at a temperature of 370C for 24 hours. Later, the bacterial colonies found on all plates were counted using a colony counter. Plates containing 30-300 colonies were used to calculate the bacterial population and recorded as CFU g-1 in each organ [8]. To determine the colony counts in CFU ml-1 of sample, the following formula was used:
Preparation of pure culture
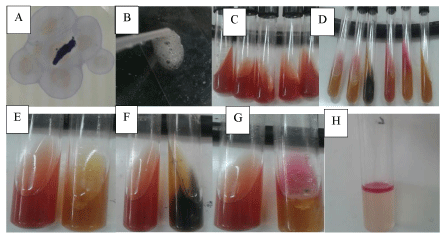
Pure cultures of the isolates were identified by using the standard procedures proposed by Barrow & Feltham [10]. In this regard, suspected colonies were picked up and re-streaked on new plates of selective medium (Mac Conkey Agar and TCBS agar). Identification of the pure culture was made by examining the colony morphology, staining characteristics, motility, oxidase activity, and oxidation-fermentation properties [9].
Morphological and biochemical characterization of isolates
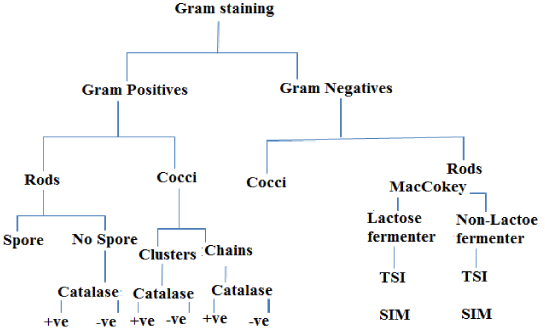
The isolates were identified using morphological and biochemical characterization following the criteria proposed in the Bergey’s Manual of Determinative Bacteriology [11]. In this regard colony morphological characterization was performed. Followed this, Gram’s staining was performed using standard procedures and observed under oil immersion objectives to determine the shape and arrangement of bacteria. Later, different biochemical characterization such as catalase, sulfide, indole tests and motility test were performed by following standard procedures as shown in Figure 2.
Antibiotic sensitivity test
The antibiotic sensitivity test was done by using the disc diffusion method. A total of seven types of antibiotic discs including Ampicillin (10µg), Streptomycin (10µg), Tetracycline (10µg), Kanamycin (5µg), Gentamycin (10µg), Erythromycin (15µg) and Amoxicillin (30µg) were used. For this, bacterial isolates were prepared with 5ml Tryptone Soy Broth (TSB) and the bacterial suspension was inoculated on a plate containing Muller-Hinton agar by swabbing. The antibiotic discs were then kept on the agar using sterile forceps and incubated at 370C for 24 hours.
Results
Prevalence and load of isolates
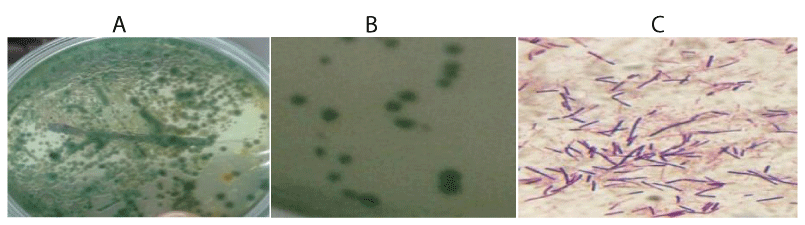
The prevalence of bacteria in the naturally infected Nile tilapia reared at the research station is shown in Tables 1,2. The results showed that 75% within the 14-17.9cm length group, 52% within the 18-21.9cm length group and 33% with the 22-26.9cm length group were infected with bacteria with a prevalence of 55%. The isolates observed in fish organs was highest in the intestine (12.43±0.55 Log 10 CFU-1g) followed by gill (12.10±0.42 Log 10 CFU-1g) and skin (10.30±1.29 Log 10 CFU-1g). and least in the liver (6.48±1.06 Log10 CFU-1g). External observation of infected fish showed clinical signs of disease (Figure 3). The results of the study showed that infection was found to be higher in gills and intestine and lower in liver and kidney.
Morphological and biochemical characteristics of bacteria
The bacteria isolated from fish tissues are presented in Table 3. The results of this study showed that the isolated bacteria belonged to the genera Vibrio, Escherichia, Aeromonas, Pseudomonas, Salmonella and Streptococcus. All the genera, except the Streptococcus, were gram negative, motile, oxidase, catalase, indole and H2S positive (Table 3 and Figures 4,5).
Antibiotic sensitivity
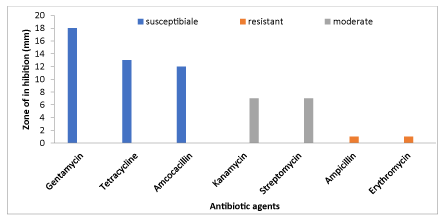
The results of the antibiotic tests of the isolates are presented in Figure 6. The results showed that gentamycin was the best antibiotic followed by Tetracycline while Ampicillin and Erythromycin were least effective.
Discussion
Intensive aquaculture production systems are carried out with high stocking densities, intensive feeding and improved management practices. In such situations, the cultured organisms are subjected to several ecological stressors, which in turn cause diseases in fish. The major diseases associated with fish are due to parasites, bacteria, viruses and fungi that reduce fish production by affecting the normal physiology of fish leading to mass mortalities. Among these, bacterial infection constitutes one of the major constraints for aquaculture that results in large-scale economic loss [12].
The occurrence of more number of pathogenic bacterial isolates found in the semi-intensive farming systems in the present study may be due to poor production management, including poor/overfeeding [13]. With different length groups, the results revealed that smaller fish were more sensitive to bacterial infection than the larger fish. This implies that as size of fish increases the prevalence of infection decreases, and this might be related to the ability of fish to withstand infections at later age. In semi-intensive and intensive aquaculture high temperature, overcrowding, occasional water restoration rate and failure to remove injured and dead fishes lead to a rise in bacterial ailments [14].
Most of the morphological and biochemical characteristics of bacterial isolates observed in the present study agreed with the reports of earlier workers [15-18]. The high bacterial count found in the intestine and gills could be associated with poor water quality and feeding practices as reported by Beveridge et al., [19], who stated that voracious feeding behavior of Nile tilapia, presence of organic matter and poor water quality of the system were responsible for the higher incidence of bacterial population in the intestine and gills of fish. The higher load of bacterial isolates in gills was also due to the fact that gills play a vital role in filtering microscopic organisms [20]. Several studies also suggested that smaller particles are entrapped by the gill filaments in a mucous leading to higher level of bacterial population [19,21,22]. Due to their broad body surface of the fish and their frequent contact with the sediment and water, skin also showed higher bacterial count. In addition, the scales could also trap detritus particles which serve as substratum for the growth of different types bacteria. On the other hand, liver showed the lowest bacterial load, which could be due to its detoxification function [22].
The present results of the antibiotic test showed that bacterial isolates were highly susceptible to gentamycin, tetracycline and amoxicillin. Isolates were moderately sensitive to Kanamycin and Streptomycin which confirm the reports of Shaw et al., [23], Sudh et al., [24] and Yu et al., [25]. The isolates found in the present study were resistant to ampicillin and erythromycin which is in agreement with the findings of others [26,27]. In conclusion, the present results clearly indicate that cultivable fishes are prone to infection by infectious and non- infectious and that it may affect fish and their product quality which leads to economic loss and livelihood of farmers who depend on small scale aquaculture. It is also a fact that commercial culture will also seriously suffer due to fish diseases as the type of culture practice followed is intensive. Hence, there is an imperative and urgent need for an integrated approach to fish health, especially general husbandry and management strategies.
- Peter RH (1993) The Ecological Implications of Body Size, Cambridge University Press. 329.
- Thiel H (1975) The size structure of deep-sea benthos. Internationale Revue der Gesamten Hydrobiologie 60: 575-606.
- Threlkeld ST (1976) Starvation and the size structure of zooplankton communities. Freshwat. Biol 6: 489-496. Link: https://bit.ly/2X4IWdR
- Kilham P, Kilham SS (1980) The evolutionary ecology of phytoplankton. The Ohysiological Ecology of Phytoplankton, L. Morris, ed 571-597. London: Blackwell Publisher.
- Semina HJ (1972) The size of Phytoplankton cells in the pacific Ocean. Internationale Revue der Gesamten Hydrobiolofie 57: 177-205. Link: https://bit.ly/2Jbl3MR
- Zohary T, Naselli-Flores L, Alster A, Fishbein T (2018) Being smaller in summer, larger. in winter: a general pattern in freshwater phytoplankton. Migal weekly seminar. in winter: a general pattern. i. in freshwater phytoplankton. Migal weekly seminar. ii. (18.11.2018) un published data.
- Ben-Tuvia A (1978) Chapter, Fishes, in: Lake Kinneret Monographiae Biologicae (C. Seruya ed), Junk Publisher 407-430.
- Bruton MN, Gophen M (1992) The Effect of Environmental Factors on the Nesting and Courtship of Tilapia zillii in Lake Kinneret, (Israel). Hydrobiologia 239: 171-178. Link: https://bit.ly/31VmgQE
- Gophen M (2017a) Experimental Study of the Feeding habits of Tilapia zillii (Gervais) in Lake Kinneret. Open Journal of Modern Hydrology 7: 1-10. Link: https://bit.ly/2FtFbIQ
- Spataru P, Viveen WJAR, Gophen M (1987) Food Composition of Clarias gariepinus (= C lazera) (Cypriniformes, Clariidae) in Lake Kinneret (Israel). Hydrobiologia 144: 77-82. Link: https://bit.ly/31UmGa3
- Drenner RW, Hambright JD, Vinyard GL, Gophen M, Pollingher U (1987) Experimental Study of Size-Selective Phytoplankton Grazing by a Filter-Feeding Cichlid and the Cichlid's Effects on Plankton Community Structure. Limnol. Oceanogr 32: 1138-1144. Link: https://bit.ly/2LhHbaI
- Gophen M (2017b) Fish-Zooplankton - A Predator-Prey Relations as a Key Factor for the Design of Zooplankton Distribution Sampling Program in Lake Kinneret, Israel. Open Journal of Modern Hydrology 7: 209-222. Link: https://bit.ly/2J73fCj
- Gophen M, Landau R (1977) Trophic Interactions Between Zooplankton and Sardine (Mirogrex terraesanctae) Populations in Lake Kinneret, Israel. Oikos 29: 166-174. Link: https://bit.ly/2NeeQVD
- Gophen M, Drenner RW, Vinyard GL (1983a) Fish Introduction into Lake Kinneret - Call For Concern. Fish Mgmt 14: 43-45. Link: https://bit.ly/2Yd7rXo
- Gophen M, Drenner RW, Vinyard GL (1983b) Cichlid Stocking and the Decline of the Galilee St. Peter's Fish (Sarotherodon galilaeus) in Lake Kinneret (Israel). Can J Fish Aquat Sci 40: 983-986. Link: https://bit.ly/2ZMhHGM
- Gophen M, Pollingher U (1985) Relationships Between Food Availability, Fish Predation and the Abundance of the Herbivorous Zooplankton Community in Lake Kinneret. Arch Hydrobiol Beih Ergebn Limnol 21: 397-405.
- Gophen M, Threlkeld ST (1989) An Experimental Study of Zooplankton Consumption by the Lake Kinneret Sardine. Archiv Hydrobiol 115: 91-95.
- Gophen M (2018) Reforming Provisional (2007-2008) Sarotherodon galilaeus Landing Decline in Lake Kinneret Isreal. Open Journal of Modern Hydrology 8: 13-27. Link: https://bit.ly/2IIwLiG
- Landau R, Gophen M, Walline P (1988) Larval Mirogrex terraesanctae (Cyprinidae) of Lake Kinneret (Israel): Growth Rate, Plankton Selectivities, Consumption Rates and Interactions with Rotifers. Hydrobiologia 169: 91-106. Link: https://bit.ly/2Yc5cDO
- Serruya CM, Gophen, Pollingher U (1980) Lake Kinneret: Carbon Flow Patterns and Ecosystem Management. Arch Hydrobiol 88: 265-302. Link: https://bit.ly/2X1XECf
- Spataru P, Gophen M (1985) Feeding Behaviour of Silver Carp (Hypophthalmichthys molitrix) (Val.) and its Impact on the Food Web in Lake Kinneret, Israel. Hydrobiologia 120: 53-61. Link: https://bit.ly/2XxLLYV
- Landau R (1975-1977) The Fish Population of Lake Kinneret; Project Report; Israel Oceanographic and Limnological Research Co. LTd. Haifa Israel 13.
- Ma H, Cui F, Liu Z, Fan Z, He W, et al. (2010) Effect of filter-feeding fish silver carp on phytoplankton species and size distribution in surface water: a field study in water works. J Environ Sci (China) 22: 161-167. Link: https://bit.ly/2Fsu0Af
- Miura T (1989) East Lake: A Phytoplanktivorous Fishes Dominated Lake Ecosystem; Otsu Hydrobiological Station, Kyoto Unuversity,No 334, 142.
- Gophen M (2014) Competitive consumption of the Lake Kinneret (Israel) plankton by Hypophthalmichthys molitrix and Sarotherodon galilaeus. Open Journal of Ecology 4: 532-542. Link: https://bit.ly/2XAEGa0
- Gophen M, Snovsky G (2015a) Silver Carp (Hypophthalmichthys molitrix,Val. 1844) Stocking in Lake Kinneret (Israel). Open Journal of Ecology 5: 343-351. Link: https://bit.ly/2KBBUeN
- Gophen M (1984) The Impact of Zooplankton Status on the Management of Lake Kinneret (Israel). Hydrobiologia 113: 249-258. Link: https://bit.ly/2IKU63A
- Gophen M (1986) Fisheries Management in Lake Kinneret (Israel). IN: Proc. Annual Meeting North American Lake Management Society, Lake Geneva, Wisconsin, USA. 327-332. Link: https://bit.ly/2X43WGw
- Gophen M (1987) Fisheries Management, Water Quality and Economic Impacts: A Case Study of Lake Kinneret. Proc. World Conference on Large Lakes, Mackinac Island, Michigan, USA. 2: 5-24.
- KLL-IOLR LKDB (1970-2013) Kinneret Limnological Laboratory, IOLR Annual Reports Lampert, W. 1987, feeding and nutrition in Daphnia. Mem.Ist Ital Idrobiol 45: 143-192.
- Lampert W (2006) Daphnia: Modelherbivore, predator and prey. Pol.J Ecol 54: 607-620. Link: https://bit.ly/2ILpLld
- Lampert W (2011) Chapter: Testing Concepts and Hypothesis, Chapter in: Daphnia: Development of a Model Organism in Ecology and Evolution. International Ecology Institute Nordbunte 23, 21385 Olendorf/Luhe Germany. 19-45. Link: https://bit.ly/2ZLv9dJ
- Gliwicz M (1980) Filtering rats, food size selection and feeding rates in cladocerans-another aspectof interspecific competition in filter-feeding zooplankton. Evolution and Ecology of Zooplankton Communities. 282-291. Link: https://bit.ly/2XwEV5X
- Dodson SI (1970) Complementary feeding niches sustained by size-selective predation. Limnol. Oceanogr 15: 131-137. Link: https://bit.ly/2FwR2Wq
- Cloen JE (2018) Why large cells dominate estuarine phytoplankton. Limnol Oceanogr 63: 392-409. Link: https://bit.ly/2NehWc3
- Maranon E, Hilligan PM, Barclela R, Gonzales N, Mourino B, et al. (2001) Patterns of Phytoplankton size structure and productivity in contrasting open-ocean environments. Inter Research MEPS 216: 43-56. Link: https://bit.ly/2X877wL
- Acevedo-Trejos EE, Maranon, Merico A (2018) Phytoplankton size diversity and ecosystem function relationships across oceanic regions. Proc Biol Sci 285. Link: https://bit.ly/2NdgerB
- Litchman E, Klaumeier CA, Yoshiyama K (2009) Contrasting size evolution in marine and freshwater diatoms. PNAS 106: 2665-2670. Link: https://bit.ly/2Y8IWe5
- Mariani P, Andersen KH, Visser AW, Barton AD, Kiorboe T (2013) Control of plankton seasonal succession by adaptive grazing. Limnbol. Oceanogr 58: 173-184. Link: https://bit.ly/2Ydi7W1
- Verdy A, Follows M, Flierl G (2009) Optimal Phytoplankton cell size in an allometric model. Inter Research MEPS 379: 1-2. Link: https://bit.ly/2RBdFOi
- Hirata T, Aiken J, Hardman-Mountford N, Smyth TJ, Barlow RG (2008) An absorption model to determine phytoplankton size classes from satellite ocean colour. Remote Sensing of Environment 112: 3153-3159. Link: https://bit.ly/2FzfjeN
- Kiorboe T (1993) Turbulence, Phytoplankton Cell Size, and the Structure of Pelagic Food Webs. Advances in Marine Biology 29: 1-72. Link: https://bit.ly/2RzIB1D
- Hansen PJ, Bjornsen PK, Hansen BW (1997) Zooplankton grazing and growth: Scaling within the 2-2, - µm body size range. Limnol. Oceanogr 42: 687-704. Link: https://bit.ly/2ZRWFqf
- Finkel ZV, Kevin JB, Quigg FA, Reesv TAV, Raven JA (2010) Phytoplankton in Changing World: cell size and elemental stichoiometry. Journal of Plankton Research 32: 119-137. Link: https://bit.ly/2Lik2oE
- Gophen M, Snovsky G (2015b) Mugilids (Mugil cepalus, Linnaeus 1758; Liza ramada, Riss 1810) Stocking in Lake Kinneret (Israel). Open Journal of Ecology 5: 389-399. Link: https://bit.ly/2J7BaL0
- Pisanti S (2005) Quasi-Cyclic fluctuationbs in St. Peter`s Fish landings in Lake Kinneret –Continues. Fisheries and Fish breeding in Israel. 1: 777-781. Link: https://bit.ly/2XvnNO9
- Serruya C (1978) Lake Kinneret, Monographiae Biologicae, Junk Publisher 32: 501.
- Walline P, Ostrovski I (1987–2016) KLDB – Kinneret Limnological Laboratory-IOLR, Annual Reports, Chapter: Fish Populations.
- Walline P, Pizanty S, Gophen M, Berman T (1993) The Ecosystem of Lake Kinneret, Israel. ICES.C.M. 1990/L: 39:1-8.
- Zohary T (2004) Changes of the Phytoplankton assemblages of Lake Kinneret after decades of predictable repetitive pattern. Freshwater Biology 49: 1355-1371. Link: https://bit.ly/31Ud2Eh

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.


 Save to Mendeley
Save to Mendeley
