International Journal of Aquaculture and Fishery Sciences
Microbial Communities Associated with the Intestinal Tract of Grey Mullets from a Mediterranean Aquatic Environment
Rosanna Floris1*, Gabriele Sanna1, Satta Cecilia Teodora1, Greta Battaggia2, Fabio De Pascale2, Alessandro Vezzi2 and Nicola Fois1
2Department of Biology, University of Padua, Via Ugo Bassi 58/B 35131 Padua, Italy
Cite this as
Floris R, Sanna G, Satta CT, Battaggia G, De Pascale Fabio, Vezzi A, et al. Microbial Communities Associated with the Intestinal Tract of Grey Mullets from a Mediterranean Aquatic Environment. Int J Aquac Fish Sci. 2024;10(1):009-019. Available from: 10.17352/2455-8400.000091Copyright License
© 2024 Floris R, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Introduction: Grey mullets comprise different species that represent the most ubiquitous teleost families in the planet’s coastal waters. They are an important proportion of the Mediterranean lagoon’s production and have been recently considered cultivated marine fish. This study aimed to explore the intestinal microbial communities of grey mullets to understand their possible ecological role for fish and the aquatic environment.
Methods: Thirty-four wild-caught mullets were sampled from a Mediterranean lagoon during four seasons and the V3-V4 hypervariable regions of 16S rRNA (Illumina MiSeq) of the fish gut were sequenced. Parameters of the aquatic environment were detected: temperature, salinity, DO, PO4, NH4, NO3, NO2, SiO, DIN, and Chla.
Results: The results indicated a high bacterial diversity (mean Shannon index: 4.74 ± 1.12; Simpson index: 0.93 ± 0.08) and variations among seasons. Sixty prokaryotic phyla were identified and the most abundant ones were: Proteobacteria (mean relative abundance 35.4% ± 17.9), Actinobacteriota (mean relative abundance 16.4% ± 9.9), and Firmicutes (mean relative abundance 10.1% ± 10.9). Bacteria belonging to the phylum Chloroflexi were relevant in autumn, Spirochaetota, Verrucomicrobiota, Fusobacteriota, and Cyanobacteria were particularly abundant in winter while Bacteroidota characterized summer fish. A total of 332 prokaryotic families were identified with 26 most abundant ones; Rhodociclaceae (Proteobacteria) were dominant in autumn, Brevinemataceae (Spirocheaetota) and Fusobacteriaceae (Fusobacteriota) were especially present in winter and the Staphylococcaceae (Firmicutes) prevailed in spring.
Conclusion: This study sheds light on the variation in the complex gut microbial community structure of Mediterranean grey mullets and their potential ecological role in protecting fish and preserving the aquatic environment.
Abbreviations
DO: Dissolved Oxygen; PO4: Reactive Phosphorus; NH4: Ammonium; NO3: Nitrate; NO2: Nitrite; SiO4: Re-active Silica; DIN: Total Dissolved Inorganic Nitrogen; Chla: Chlorophyll a
Introduction
The interest in the intestinal microbiome of aquatic animals is getting more and more pivotal because its knowledge can contribute to the development of effective strategies for fish rearing in captivity, by manipulating gastrointestinal (GI) microbial communities to promote health and productivity through novel therapeutics and feed additives [1,2]. Moreover, flexibility in the gut microbiome may play a role in biological diversity conservation, enabling fish to colonize new and different aquatic environments [3]. The fish GI tract is a complex ecosystem composed of a dynamic consortium of microorganisms that play critical roles in nutrition, energy sources, and host health to reduce or inhibit pathogenic microbes and for the safety of the environment [4]. Fish gut microflora can be divided into two groups: the resident (autochthonous) and the transient (allochthonous) communities [5]. The resident can adhere to and colonize the mucosal surfaces, or occur within the epithelial tissues, while the transient communities are characterized by non-adherent free-living microorganisms although they inhabit microniches, especially during periods of stress [6]. The intestinal microbiota of aquatic animals has higher fluidity than terrestrial animals and changes in various factors such as temperature, salinity, and trophic level [7-10]. The studies on Mediterranean fish species indicated that it is influenced by species, sex, age [11], and physiology other than epigenetic factors (season, feeding regimen, water temperature, and salinity) [12,13]. However, the existence of a core gut microbiota within and between different species independent of diet and geographic location does exist [14]. Thus, the monitoring of bacteria present in healthy wild fish in their natural environment is the first step for use in captivity [15]. Research on intestinal bacteria of a wide range of fish species has mostly reported on the isolation, identification, and evaluation of cultivable bacteria; however, only a small part (< 2%) of the GI microbiota may be cultured and these types of procedures fail to provide information of the microbial community as a whole. Metabarcoding sequencing by various Next-Generation Sequencing (NGS) platforms has emerged as a method to discover new groups of micro-organisms with greater accuracy in environmental systems in spatial and temporal scales [5,10,16]. Illumina MiSeq (Illumina, USA) has been widely used for 16S rRNA gene sequencing of gastrointestinal tract microbiota of freshwater [17] and marine fish [16,18].
Fish belonging to the Mugilidae family, commonly known as the grey mullets group, comprise a great number of species and they are one of the most ubiquitous teleost families in the planet’s coastal waters [19]. They have omnivorous and herbivorous feeding habits [20] and have been recently considered a cultivated marine fish that can be fed with “alternative” energy such as insects and cost-effective materials, plants, production dis-cards, etc., contributing to the realization of the goal of sustainability in aquaculture [21]. In this sense, grey mullets represent an interesting resource for aquaculture use although large-scale production faces numerous challenges represented by the ecological carrying capacity of existing sites and environmental impact [22]. In any case, the valorization of fish species less considered for the market is a way to preserve the more valued ones and the biodiversity of the aquatic environment [23].
Among Mediterranean Mugilidae, Mugil cephalus (Linnaeus, 1758), Chelon ramada (Risso, 1827), Chelon labrosus (Risso, 1827), Chelon saliens (Risso, 1810) and Chelon auratus (Risso, 1810) are some of the most representative fish species in Sardinian lagoons [22,24]. Above all, flathead grey mullet, M. cephalus, represents about 50% of world mullet production [19], possesses high economic value, and is appreciated in the food market for its eggs processed to obtain seafood which is known by different names such as Avgo-taracho (Grece), Karasumi (Japan) or Bottarga (Italy), depending on the geographical production area [25,26]. In Italy, grey mullets represent an important proportion of the production of coastal lagoons [27] and their culture is carried out in extensive systems, based on natural cycles and dynamics [28]. Production is based on wild fry availability and it cannot compete with intensive cage culture at sea, but it aims to combine environmental compatibility with economic sustainability. This is an advanced form of coastal lagoon management and represents one of the most interesting examples of coastal lagoon management in the world [29]. According to official sources in 2015, the Italian organic mullet species (M. cephalus, C. aurata, C. saliens, and C. labrosus) production is estimated at 80 tonnes (source EUMOFA).
Earlier studies reported the composition and the predicted functions of the associated gut microbiota on M. cephalus of different ages from Northwest Pacific marine environments [30], from Chinese coastal marine areas [31], and on Mediterranean wild thick–lipped grey mullets (C. labrosus) [23]. In this work, a metabarcoding study on 34 wild mullets from a Mediterranean transitional aquatic environment has been carried out to analyze, as a first step, the structure of their intestinal bacterial communities during different seasons. These results shed light on the variation in the complex gut microbial community structure of Mediterranean grey mullets and its potential biotechnological role for fish and the aquatic environment.
Materials and methods
Study area and fish samplings
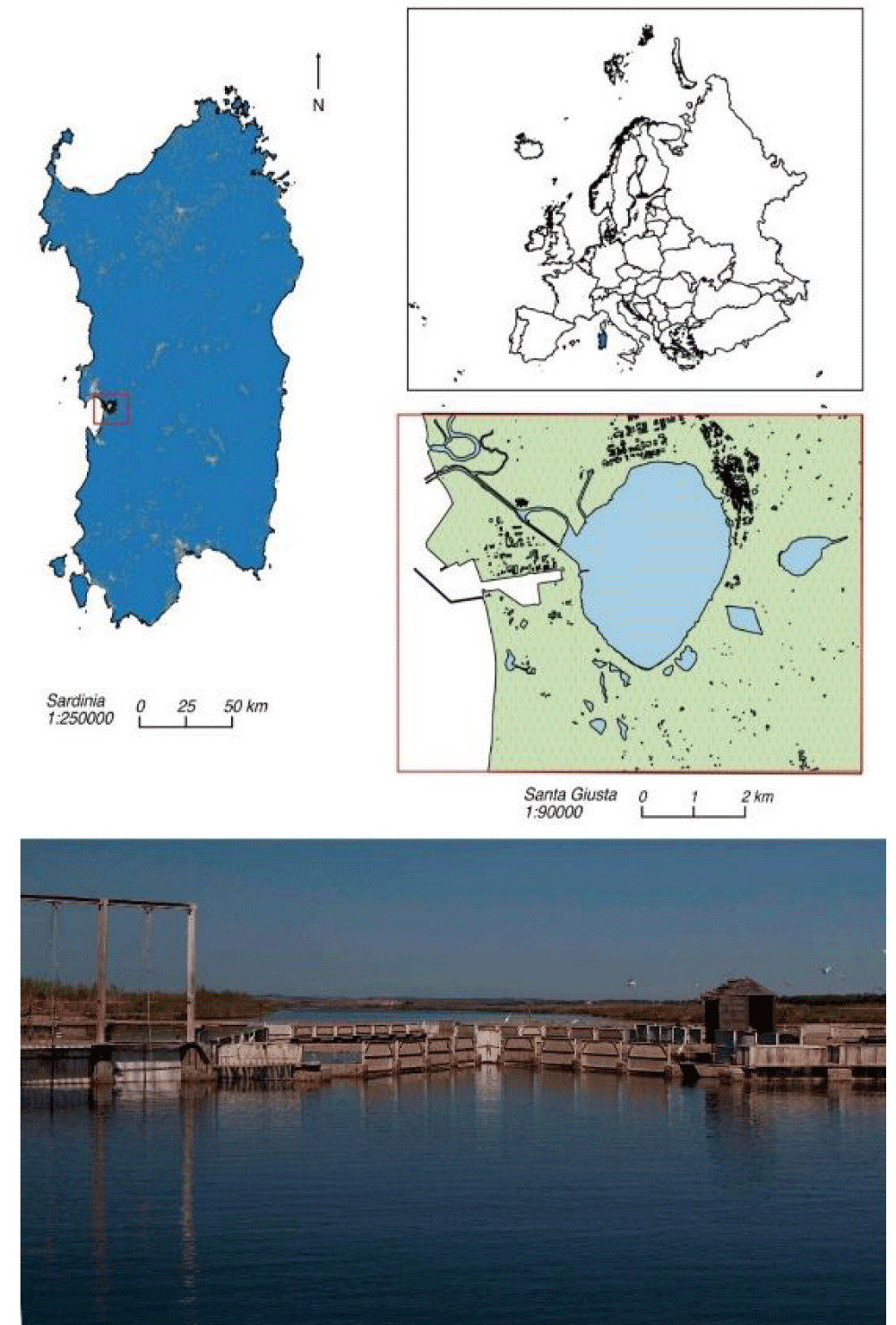
Thirty-four wild-caught mullets, destined for the local food market, were captured by professional fisheries on September 27th, 2018 (autumn), February 10th, 2019 (winter), July 10th, 2019 (summer), and May 18th, 2021 (spring) from Santa Giusta Lagoon (Central west coast of Sardinia (Italy) (coordinates: Lat 39°52’N, Long 8°35’E). The fish were transported inside a refrigerated bag to the Agris Bonassai laboratory within 3-4 hours. Species were determined on a morphological basis, according to identification keys [32,33].
The aquatic area (8.6 km2 and 1.0 m mean depth) is a research site of the “Marine Ecosystems of Sardinia” of the Italian Long-Term Ecological Research network (www.lteritalia.it; https://deims.org/6f7581f0-e663-4681-bf9d-4668d6c3f2ba), recognized as a Site of Community Importance for European Union (SCI ITB030037) and is designated by the Sardinian Government as a protected area for animals (INFS code: OR0211) (Figure 1).
Fish measurements and preparation of gut samples
Body and gut weight and total length were measured. The intestine (mean weight 14 ± 6 g) was aseptically removed, diluted (10%, w/v) in saline solution (0.90% NaCl), and homogenized in plastic bags by Stomacher® 400 (FermionX Ltd, Worthing, UK) at room temperature. The whole intestines of fish were collected. The samples for microbiological analyses (homogenates) were made up by mixing the guts of two-three individuals of the same species and immediately stored at -80 °C until DNA extraction. The pooling of the samples was performed to minimize the individual variations in the fish [34,35]. It was not possible to find mullets of the same species in the local food market during the considered seasons. This was probably due to the migratory behavior of all the wild species that enter and go out from the lagoon and the management of the lagoon (the aquatic area is submitted to different closures to the sea by the fishermen for production purposes) and other uncontrolled factors (the great environmental instability of the lagoon).
Environmental characterization
To explore the seasonal dynamics in the lagoon during the study period, a total of 49 water samples were collected. Water samplings were carried out monthly in autumn 2018, winter and summer 2019, and in May 2021. Temperature, salinity, and Dissolved Oxygen (DO) were measured in situ using a YSI 6600 v2 (YSI Inc., Yellow Springs, USA) multi-parameter probe.
Samples for nutrient analyses were collected at about 30 cm depth. Nutrients were analyzed within a few hours after sampling. Concentrations of inorganic nutrients such as reactive Phosphorus (PO4), Ammonium (NH4), Nitrate (NO3), Nitrite (NO2), and reactive Silica (SiO4) were determined on the filtered samples according to Strickland and Parsons, 1972 [36]. Total Dissolved Inorganic Nitrogen (DIN) was calculated as the sum of NH4, NO3, and NO2. Chlorophyll a (Chla) was determined following the SCOR-UNESCO protocol 1997.
The Dipartimento Specialistico Regionale Idrometeoclimatico (SAR-ARPAS: http://www.sar.sardegna.it/) provided daily data on rainfall (Rain).
Rain data were obtained by summing daily rainfall values to get seasonal accumulations.
DNA extraction, amplification, and sequencing
DNA was extracted from ca. 0.750 g of homogenate (n = 12) obtained by mixing the guts of three individuals of the same species.
All extractions were performed according to the PowerSoil DNA Isolation Kit (Qi-agen®, Hilden, Germany) following the manufacturer’s instructions. DNA quality and quantity were assessed by spectrophotometry (NanoDrop®, Wilmington, DE, USA) and fluorometrically (Qubit® Life Technologies, Paisley, UK) to ensure optimal measurement of DNA quantity and purity. The V3-V4 region of the 16S rRNA gene was amplified using universal bacterial primer pair 341F (5’-CCTACGGGNBGCASCAG-3’) and 785R (5’-GACTACNVGGGTATCTAATCC-3’). The PCR mixtures, in a final volume of 25 µl, were as follows: 10 ng of template DNA, 0.5 U of Phusion High-Fidelity DNA polymerase (Thermo Fisher Scientific, Waltham, USA), 1X Phusion HF buffer, 0.5 µM of each primer and 200 µM of each dNTP. PCRs (98 °C for 4 min; 35 cycles of 98 °C for 20 s, 57 °C for 30 s, 72 °C for 30 s; 72 °C for 5 min) were set up in triplicate to smooth possible in-tra-sample variance. PCR products were visualized on 1.5% agarose gels, then amplicon triplicates were pooled and purified using 0.8X volumes of AMPure XP beads (Beckman Coulter, Brea, CA, USA).
The pooled PCR products were indexed and subsequently normalized according to the “16S Metagenomic Sequencing Library Preparation” protocol, with minor adjustments: 1) 0.5 U of Phusion High-Fidelity DNA polymerase (Thermo Fisher Scientific, Waltham, USA) was used for each reaction and 2) PCR amplicons were purified using 0.7X volumes of AMPure XP beads (Beckman Coulter, Brea, CA, USA). Finally, amplicon libraries were equally pooled and sequenced using the Illumina MiSeq system, in the 2 x 300 bp format (Illumina, San Diego, CA, USA). The 16S amplicon sequences generated for this study can be found in the Sequence Reads Archive (SRA) at NCBI under the accession number PRJNA893889.
Bioinformatic analyses
Raw reads were initially processed with Cutadapt, v. 2.1 to remove primer sequences and reads shorter than 100 bp [37]. All further analyses were conducted in R, v. 4.1 (R CoreTeam, 2019). At first, using DADA2, v. 1.20 [38], forward reads were trimmed at 270 bases and reverse reads at 170 bases, also truncating reads where bases with quality 2 were found and allowing 0 Ns and maximum expected errors equal to 2. At the end of this process, reads shorter than 20bp were discarded. The resulting reads were then denoised and merged to obtain the Amplicon Sequence Variants (ASVs) in the samples. Taxonomy annotation was performed in DADA2 using the SILVA database v. 138 clustered at 99% identity [39]. Subsequent analyses were performed using the phyloseq R package [40] for data handling and further filtering. All ASVs assigned to Eukarya, mitochondria, and chloroplast, or not assigned at the phylum level, were removed; only ASVs with total counts above 10 reads across all the samples were retained for further analyses. Rarefaction curves were produced with a vegan R package. Alpha diversity indexes were computed with phyloseq dedicated functions; beta diversities principal coordinates analysis plots were computed on Bray-Curtis distances matrixes. To describe the different microbial communities, their differences, and similarities, relative abundances of ASVs were computed in all samples. A Venn diagram was drawn to reveal the unique and shared families in the different samples. ASVs were grouped at family levels and analyzed with the Vienna package.
Results
Fish biometry
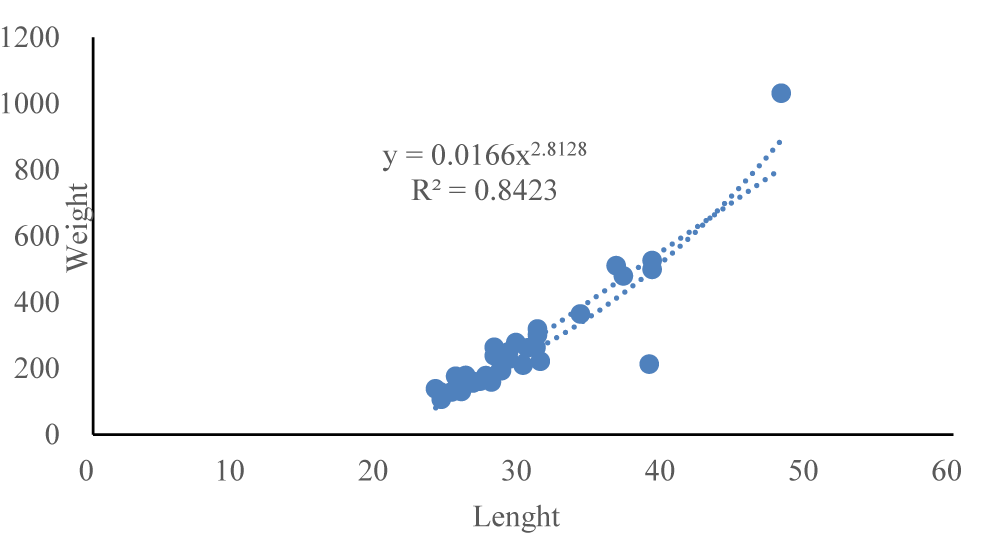
The morphological traits of the wild mullets are represented by the equation shown in Figure 2.
The graphic shows the relationship between total weight and length of the mullets and the determination coefficient (R2) indicates a good correlation (r = 0.916) between these biometric variables.
Aquatic environmental characterization
Physical and chemical parameters of the aquatic environment during the period of study are reported in Table 1. Seasonal rain accumulates showed the highest mean values (396.2 mm) in autumn 2018 and the lowest ones (40 mm) in summer 2019. Mean temperature data showed seasonal variations with the highest values recorded in summer 2019 (26.83 ± 2.03 °C) and the lowest ones in winter 2019 (12.59 ± 2.19 °C). Mean salinity data indicated an increase along the seasons from autumn 2018 (20.84 psu ± 7.54) to summer 2019 (35.10 psu ± 2.99). The lowest DO (5.78 mgL-1 ± 0.67) mean values were registered in summer 2019 while the highest (10.03 mgL-1 ± 1.07) ones were in winter. Regarding nutrients, the highest mean values were detected in autumn for all those considered (Table 1). The lowest mean chlorophyll content (0.55 mg m-3 ± 0.26) was detected in spring while and the highest mean value was in autumn (22.42 mg m-3 ± 4.84).
16S rDNA sequencing
Diversity analysis: 16S barcode sequencing yielded a total of 4,995,563 row sequences. After the initial steps of filtering, denoising, merging, and chimera removal, a total of 2,618,159 sequences were obtained. From these, 12,045 Amplicon Sequence Variants (ASVs) were identified considering only those that were not assigned to eukaryotic phyla. Considering all samples, ASVs with fewer than 10 sequences were discarded, resulting in a final number of 7817 ASVs. The rarefaction curves showed that the sequencing effort was sufficient to assess the biodiversity of the samples (Supplementary Material. Figure S1). Table 2 shows the number of total counts relative to the ASVs remaining after the different filtration steps, the number of ASVs identified in the microbiota of each fish intestinal sample in each season, and the microbial α-diversity expressed by Shannon and Simpson indexes. No significant difference was found among the α-diversity indexes of samples.
Composition of intestinal microbiota of grey mullets
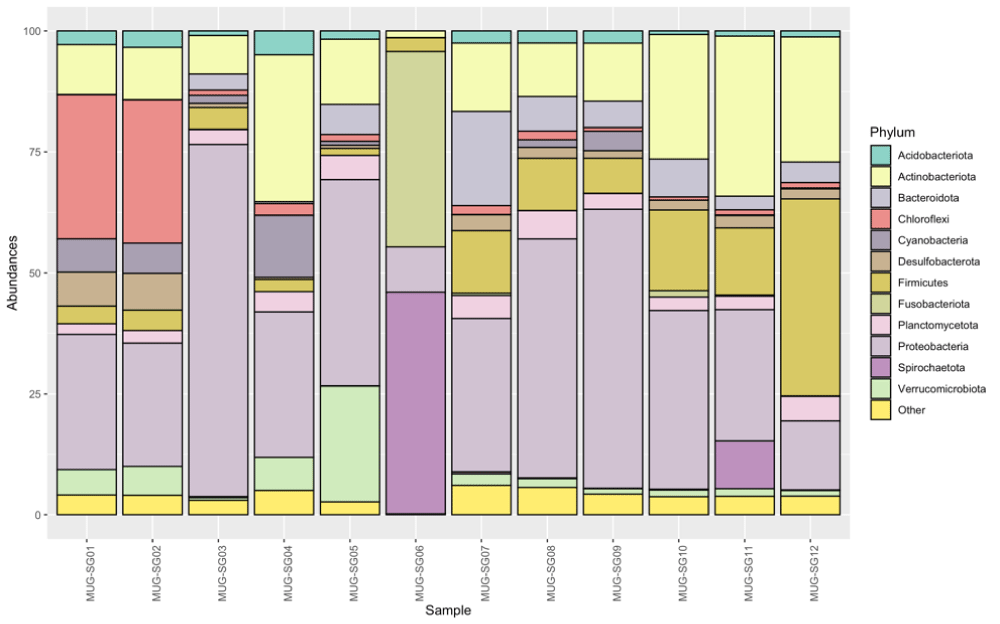
Bacterial phyla: The 7817 ASVs identified in the dataset were assigned to 60 different phyla (Supplementary Table 1). Figure 3 shows the relative abundances of the phyla identified in the mullets during the various seasons. Phyla with a median abundance of more than 1% or a variance greater than 85% of the entire dataset were considered in the analyses. In this regard, Acidobacteriota, Actinobacteriota, Bacteriodota, Chloroflexi, Cyanobacteria, Desulfobacterota, Firmicutes, Planctomycetota, Proteobacteria and Verrucomicrobiota had a median abundance of more than 1%, while Spirochaetota and Fusobacteriota showed an overall low abundance but high variability among the samples, as they both accounted for more than 40% of the prokaryotic community in MUG-SG06 sample during winter. All the remaining 48 phyla were included in the “Other” group (Figure 3 and Supplementary Table 2).
The gut microbiota of grey mullets was dominated by Proteobacteria (35.4% ± 17.9), followed by Actinobacteriota (16.4% ± 9.9) and Firmicutes (10.1% ± 10.9) which together accounted for 61.9% of the total population identified, and constituted the “core” microbial group. In this “core” microbiome, the Actinobacteriota resulted significantly higher in fish captured in May (spring) (84.68%) with respect to February (winter) (43.82%), July (summer) (37.21%) and September (autumn) (29.8%) (p < 0.01) and the Firmicutes were at the highest number in spring, but they were significantly higher in summer with respect to winter and autumn and also in autumn with respect to winter (p < 0.05) (Figure 4). Generally, the highest number of bacterial phyla was observed in summer; however, the few phyla that appeared specific to a given season, always showed low abundances, so they should not have a strong effect on the structure and dynamics of the microbial communities in the fish gut (Supplementary Table 1).
To explore the prokaryotic diversity of a single fish intestinal sample, the three phyla with the highest abundance were examined in each of them (Supplementary Table 2). These identified phyla were represented by Actinobacteriota, Bacteriodota, Chloroflexi, Cyanobacteria, Firmicutes, Fusobacteriota, Proteobacteria, Spirochaetota and Verrucomicrobiota. On average, the top three phyla identified in each sample comprised more than 75% of the total abundance and this means that these phyla accounted for most of the microbial community in the gut of each sample. The only exceptions were MUG-SG01, MUG-SG02, and MUG-SG07 in which, however, the most abundant three phyla represented more than 65% of the total abundances.
Considering the gut microbiome during the different periods of study, the ASVs belonging to the Chloroflexi phylum were particularly relevant for fish captured in September (autumn) (MUG-SG01 and MUG-SG02), while much lower abundances of this phylum were found in all the other samples captured in the other seasons (Supplementary Table 1 and Figure 4). Remarkably, the C. labrosus samples caught in February (winter) (MUG-SG05 and MUG-SG06) showed marked differences with respect to each other. In particular, fewer ASVs were identified in the MUG-SG06 sample, with a large abundance of Spirochaetota (45.8%) and Fusobacteriota (40.4%) as already highlighted, which were at very low abundance or even absent in all the other samples (Supplementary Table 1).
Bacterial families
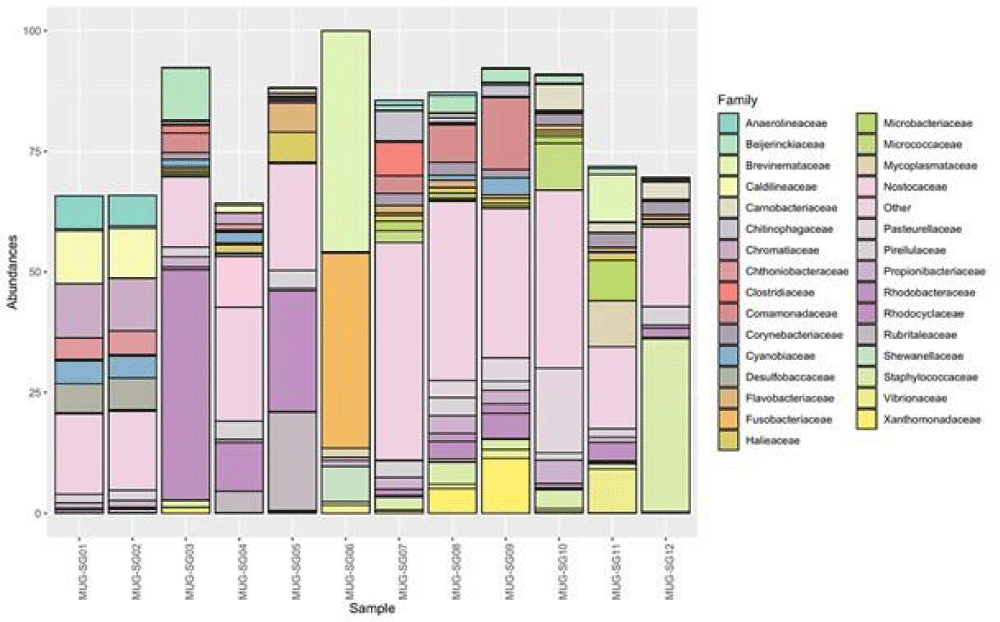
Looking at deeper taxonomy levels, a total of 332 families were identified. Not all the identified ASVs were assigned to known families and were therefore removed from the analysis. In this regard, 70.1% of the ASVs, comprising 84% of the reads, remained. However, most of the known families had total abundances in all samples below 1%, meaning that they had a low impact on the overall microbial communities. On the other hand, 88 families overpass this threshold (Supplementary Table 3). To have a broad picture of the identified families and their impact on the microbial communities of the mullets’ gut, their abundances were further investigated. Figure 5 shows the relative abundances of all the identified families, highlighting those having median abundance in the samples higher than 1% or variance higher than 75% of the entire dataset. All remaining families have been enclosed in the “Other” group, which reaches a large fraction of the overall abundance in some samples. However, some families were found to have a large impact in terms of abundance on the communities, as discussed below.
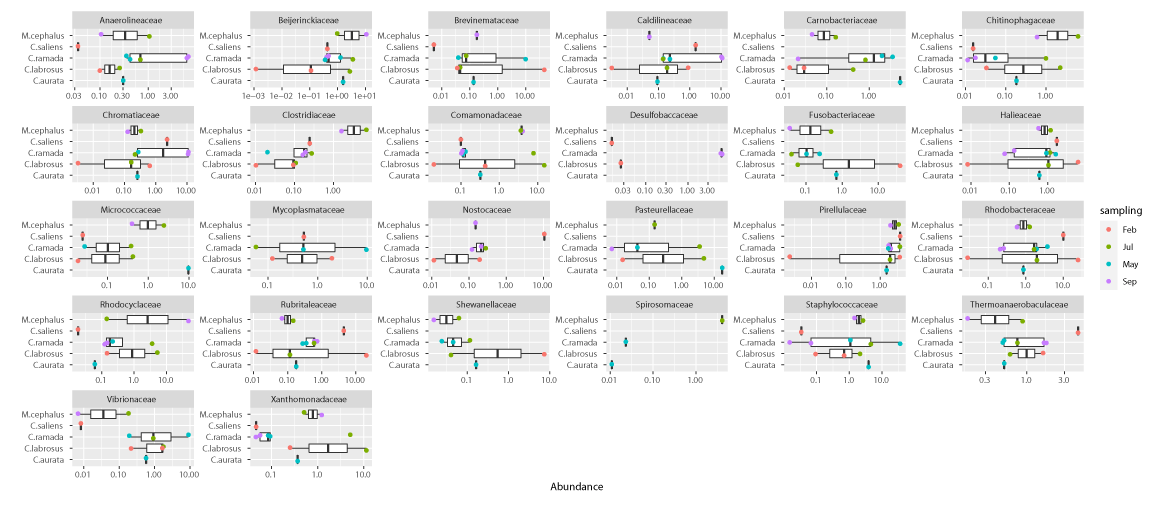
Sixty microbial families were shared among all fish and represent different amounts of the overall microbial communities ranging from 37.8% to 81.8% with a median value of 54.6% (Supplementary Table 3). Eleven known families were found to be ubiquitous in all fish and seasons. These families were Staphylococcaceae, Comamonadaceae, Beijerinckiaceae, Propionibacteriaceae, Rhodobacteraceae, Rubritaleaceae, Ilumatobacteraceae, DEV007, Halieaceae, Pirellulaceae, and Desulfocapsaceae (Supplementary Table 3). These families had average abundances of less than 5% in all gut microbial communities analyzed. Nevertheless, some trends could be highlighted in certain seasons, such as Staphylococcaceae in May (spring), which reached 35.8% abundance; Rhodobacteraceae and Rubritaleaceae in February (winter), exceeding 20% abundance; Comamonadaceae in July (summer) had an abundance higher than 14%; and Beijerinckiaceae in September (autumn), which had around 10% of the counts (Supplementary Table 3 and Figure 5). In most other cases abundances were lower than 1.9%. A total of twenty-six most abundant bacterial families were found. Looking at the three most abundant families within each intestinal microbial community of the different mullet samples, on average, these three families represented 38.4% of the total abundance, even if they showed very high variability (st. dev: 21.9) (Figure 6). This is because, in half of the samples, the three most abundant families do not reach 30% of the total abundance while in some others they largely exceed 50% (Supplementary Table 4).
The seasonal distribution of intestinal bacterial families shows that Rhodociclaceae (Proteobacteria) (47.7%) was the most abundant group in September (autumn), Brevinemataceae (Spirocheaetota) (45.8%), and Fusobacteriaceae (Fusobacteriota) (40.4%) were dominant in February (winter), and Staphylococcaceae (Firmicutes) (35.8%) prevails in May (spring). On the other hand, the relative abundance of the first three families in the fish caught in July (summer) did not appear particularly high, as only Comamonadaceae and Xanthomonadaceae (both Proteobacteria, respectively 14.92% and 11.39%) exceeded 10% abundance in MUG-SG09 (Supplementary Table 4). The other less abundant families which characterized each season were: Chromatiaceae (Proteobacteria), Caldilineaceae (Chloroflexi), Beijerinckiaceae (Proteobacteria), Anaerolineaceae (Chloroflexi) and Desulfobaccaceae (Desulfobacterota) in September (autumn); Rubritaleaceae (Verrucomicrobiota), Rhodobacteraceae (Proteobacteria), Nostocaceae (Cyanobacteria), Shewanellaceae (Proteobacteria), Halieaceae (Proteobacteria), Thermoanaerobaculaceae (Acidobacteriota) in February (winter); Xanthomonadaceae (Proteobacteria), Clostridiaceae (Firmicutes), Chitinophagaceae (Bacteroidota), Spirosomaceae (Bacteroidota) identified in July (summer), and Pasteurellaceae (Proteobacteria), Brevinemataceae (Spirochaetota), Micrococcaceae (Actinobacteriota), Mycoplasmataceae (Firmicutes), Vibrionaceae (Proteobacteria), Carnobacteriaceae (Firmicutes), Pirellulaceae (Planctomycetota) observed in May (spring) (Supplementary Table 3).
Discussion
In this study next-generation sequencing technologies (NGS) and bioinformatics analysis allowed us to gain a greater knowledge of the microbial communities (both the resident and the transient) associated with the gut of Mediterranean wild grey mullets in response to a variety of environmental aquatic factors. The study revealed the structure of a complex ecosystem is, highly influenced by the aquatic environment during different seasons. The fish specimens analyzed represent the typical edible wild fauna from Sardinian coastal lagoons and other Mediterranean transitional aquatic environments [27]. Research attention has been focused on the aquaculture of M. cephalus, which represents a traditionally harvested and consumed fish in various European countries such as Italy, Spain, and France, especially appreciated in Tunisia, Egypt, and Taiwan, and a suitable species for feeding populations in developing countries [20,26]. Moreover, the culture of grey mullets Mugilidae species is considered a priority within the current strategies of sustainable European aquaculture [21]. To the authors’ knowledge, various studies were made on the gut microbiota of cultured [41] and wild mullets [23,31] and the present study has provided more information on the biodiversity of mullets intestinal microflora and their biotechnological potential. Previous studies on Mediterranean grey mullets have shown some interesting biotechnological traits of intestinal cultivable bacteria as a source of bioactive compounds with immunological and bioremediation functions [42].
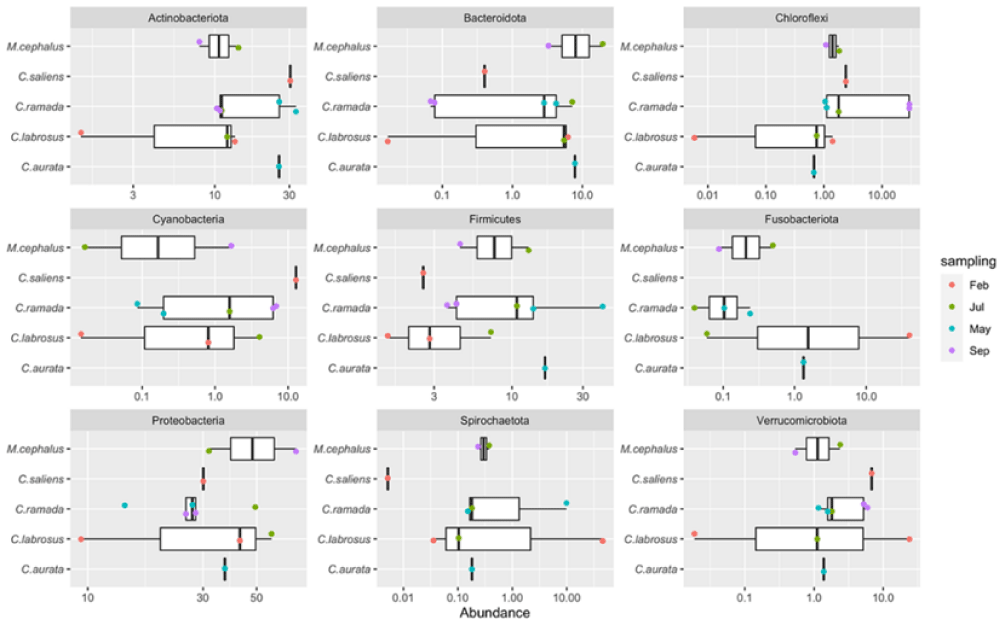
The outlined microbiome confirms that the acquisition and maintenance of the gut microbiota is a very complex process, which is dictated by both environmental factors and host physiological pressures [43,44]. Indeed, each captured grey mullet of this study is characterized by a specific breeding period that determines the migratory behavior that depends on spawning, endocrine mechanisms, photoperiod, temperature, and feeding activity [45]. This work has revealed the presence in the mullets of dominant main phyla: Gram-negative (Proteobacteria) and Gram-positive (Actinobacteriota and Firmicutes) which accounted for 61.9% of the total prokaryotic population identified across all the intestinal samples and constitute the “core” microbial community in the mullets’ gut. This suggests the potential role of these core taxonomic groups for vital functions in the nutrition and/or the immunity of the fish. From an ecological point of view, it was interesting to observe a seasonal influence on the gut microbial composition with a dominance, in spring, of Actinobacteria and Firmicutes which were also detected in the intestinal microflora of other marine and freshwater fish [14,46-48].
The present study detected the occurrence of the phylum Chloroflexi, which was particularly abundant in the C. ramada individuals caught in autumn (both samples were above 29.5%). Other papers reported the presence of Chloroflexi in the microbial communities of M. cephalus gut [30] and in a wide range of aerobic and anaerobic habitats including sediments, hot springs, and methanogenic reactors, where these bacteria are supposed to have a role in sludge stabilization and breakdown [49]. These authors reported that bacteria of Chloroflexi phylum, commonly isolated from sludge matrices, have a role in bioremediation processes, being able to degrade complex polymeric organic compounds to low molecular weight substrates (sludge granulation). Moreover, Liang, et al. [50] described Chloroflexi as a component of bacterial communities from petroleum reservoirs and its involvement in toluene degradation. In particular, members of the Chloroflexi phylum belonging to the Anaerolineaceae family found at an abundance of more than 6% in the mullets collected in autumn, are described in the literature as methanogenic bacteria (hydrocarbon degrading), frequently encountered in the presence of petroleum. The presence of bacteria able to degrade toxic substances as aromatic compounds (styrene and fluorobenzoate) on Chelon labrosus was also pointed out using PICRUSt functional analysis [23], although their role is still unclear and most of this group of microorganisms remains uncultured, and understudied [51].
In this study, the most abundant bacterial family was represented by Rhodocyclaceae (47.7%) (beta-Proteobacteria) as observed by Le and Wang [30] in the gut of M. cephalus from the Taiwan Strait. Interestingly, the Rhodocyclaceae species were described by different authors for producing bioactive metabolites and, in particular, being able to transform perchlorate into harmless chloride [52-54]. In this regard, Guarino, et al. [54] reported about the genera Azospira and Dechloromonas of the Rhodocyclaceae family, able to transform perchlorate into harmless chloride, which is widely distributed in different environments such as soil and groundwater. Nowadays, perchlorate (ClO4-) is a ubiquitous ion released into the environment by anthropogenic activity although significant quantities of perchlorate are naturally formed in the atmosphere, especially during thunderstorms [53,55,56]. The main effect on human beings is its action on the thyroid gland by inhibiting iodide uptake and synthesis of thyroid-stimulating hormone, with serious impairments of growth, metabolism, and reproduction. Another dominant bacterial family identified in the mullets is the Brevinemataceae (Spirocheaetota). This microbial group was found to be very abundant in winter (45.8%) as also reported by Le and Wang [30] for the gut of M.cephalus and by García-Márquez, et al. [23] for C. labrosus individuals. Members of the Brevinemataceae family are described by other authors as producing butyrate [57] which may have an intestinal barrier function and support mucosal immunity [58]. Throughout this study, another family represented by Staphylococcaceae (Firmicutes) was detected significantly in the gut of fish sampled in spring (35.8%). Different papers described several biotechnological activities of Staphylococcaceae [59] and its capacity for degrading hydrocarbon [50]. Generally, the Firmicutes phylum is regarded as beneficial bacteria to the host since it comprises the group of lactic acid bacteria that are highly studied for their probiotic properties [60,61]. In this regard, the Lactobacillales order was identified with an abundance of more than 1% in all fish species caught in spring and summer, seasons with environmental conditions more suitable for this type of bacteria.
The present work is a first study via 16S rRNA metabarcoding technology on the intestinal communities of different species of Mediterranean grey mullets from a Sardinian aquatic environment; as already remarked, it was impossible to find mullets of the same species in the local food market during the different seasons of study, probably due to the different migratory behavior of these wild species and the management of the lagoon. Other metagenetic studies will be carried out considering the single fish species along different seasons and from other Mediterranean aquatic environments to have a broader view of the ecology of the bacterial communities associated with the intestinal tract of wild grey mullets.
Conclusion
The findings of the present work provide interesting insights into the diversity and biotechnological potential of the symbiotic intestinal communities hosted by Mediterranean grey mullets.
The results add new important insights into the intestinal microbial ecology of these fish as alternative candidate species and for rational use in aquaculture, following the EU policy for innovative technology and environmentally and commercially sustainable for a rational use.
Future research could be driven to the mullet aquaculture for preserving their intestinal microbiome which is an added value to protect fish and to preserve the aquatic habitat, a prerequisite of a sustainable aquaculture.
The authors would like to thank Dr. Riccardo Diciotti and Dr. Jacopo Culurgioni for collecting grey mullet samples and sharing their expertise in the identification of fish; and the Santa Giusta Cooperative for providing fish.
- Hai NV. The use of probiotics in aquaculture. J Appl Microbiol. 2015;119:917-935. Available from: https://pubmed.ncbi.nlm.nih.gov/26119489/
- Dittmann KK, Rasmussen BB, Castex M, Gram L, Bentzon-Tilia M. The aquaculture microbiome at the centre of business creation. Microb Biotechnol. 2017;10:1279-1282. Available from: https://pubmed.ncbi.nlm.nih.gov/29064164/
- Jones J, DiBattista JD, Stat M, Bunce M, Boyce MC, et al. The microbiome of the gastrointestinal tract of a range-shifting marine herbivorous fish. Front Microbiol. 2018;9:1-13. Available from: https://pubmed.ncbi.nlm.nih.gov/30210475/
- Wang AR, Chao R, Ringø E, Zhou ZG. Progress in fish gastrointestinal microbiota research. Rev Aquac. 2018;10:626-640. Available from: https://onlinelibrary.wiley.com/doi/10.1111/raq.12191
- Legrand TPRA, Wynne JW, Weyrich LS, Oxley APA. A microbial sea of possibilities: current knowledge and prospects for an improved understanding of the fish microbiome. Rev Aquac. 2020;12:1101-1134. Available from: https://doi.org/10.1111/raq.12375
- Banerjee G, Ray AK. Bacterial symbiosis in the fish gut and its role in health and metabolism. Symbiosis. 2017;72:1-11. Available from: https://doi.org/10.1007/s13199-016-0441-8
- Denev S, Staykov Y, Moutafchieva R, Beev G. Microbial ecology of the gastrointestinal tract of fish and the potential application of probiotics and prebiotics in finfish aquaculture. Int Aquat Res. 2009;1:1-29. Available from: https://journals.iau.ir/article_673235_fc3c20524391059180cdbf20d0193190.pdf
- Guerreiro I, Serra CR, Enes P, Couto A, Salvador A, Costas B, et al. Effect of short chain fructooligosaccharides (scFOS) on immunological status and gut microbiota of gilthead sea bream (Sparus aurata) reared at two temperatures. Fish Shellfish Immunol. 2016;49:122-131. Available from: https://pubmed.ncbi.nlm.nih.gov/26721230/
- Ringø E, Zhou Z, Vecino JL, Wadsworth S, Romero J, Krogdahl Å, et al. Effect of dietary components on the gut microbiota of aquatic animals: a never-ending story? Aquac Nutr. 2016;22:219-282. Available from: https://doi.org/10.1111/anu.12346
- Sullam KE, Essinger SD, Lozupone CA, O'Connor MP, Rosen GL, et al. Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol Ecol. 2012;21(13):363-3378. Available from: https://pubmed.ncbi.nlm.nih.gov/22486918/
- Piazzon MC, Naya Català F, Simó Mirabet P, Picard-Sánchez A, Roig FJ, Knight R, et al. Sex, age, and bacteria: how the intestinal microbiota is modulated in a protandrous hermaphrodite fish. Front Microbiol. 2019;10:2512. doi:10.3389/fmicb.2019.02512
- Floris R, Manca S, Fois N. Microbial ecology of the intestinal tract of gilthead sea bream (Sparus aurata Linnaeus, 1758) from two coastal lagoons of Sardinia (Italy). Trans Water Bull. 2013;7:4-12. Available from: http://siba-ese.unisalento.it/index.php/twb/article/view/13419
- Floris R, Sanna G, Satta C, Piga C, Sanna F, Lugliè A, et al. Intestinal microbial ecology and fillet metal chemistry of wild grey mullets reflect the variability of the aquatic environment in a western Mediterranean coastal lagoon (Santa Giusta, Sardinia, Italy). Water. 2021;13:879. doi:10.3390/w13060879
- Nikouli E, Meziti A, Antonopoulou E, Mente E, Kormas KA. Gut bacterial communities in geographically distant populations of farmed sea bream (Sparus aurata) and sea bass (Dicentrarchus labrax). Microorganisms. 2018;6:92. Available from: https://pubmed.ncbi.nlm.nih.gov/30200504/
- Tarnecki AM, Patterson WF, Arias CR. Microbiota of wild-caught red snapper Lutjanus campechanus. BMC Microbiol. 2016;16:245. Available from: https://pubmed.ncbi.nlm.nih.gov/27769187/
- Yukgehnaish K, Kumar P, Sivachandran P, Marimuthu K, Arshad A, Bilal Ahmad Paray, et al. Gut microbiota metagenomics in aquaculture: factors influencing gut microbiome and its physiological role in fish. Rev Aquac. 2020;12:1903-1927. doi:10.1111/raq.12416
- Kashinskaya EN, Belkova NL, Izvekova GI, Simonov EP, Andree KB, et al. A comparative study on microbiota from the intestine of Prussian carp (Carassius gibelio) and their aquatic environmental compartments, using different molecular techniques. J Appl Microbiol. 2015;119:948-961. Available from: https://pubmed.ncbi.nlm.nih.gov/26189342/.=
- Dehler CE, Secombes CJ, Martin Samuel AM. Environmental and physiological factors shape the gut microbiota of Atlantic salmon parr (Salmo salar L.). Aquaculture. 2017;467:149-157. Available from: https://www.sciencedirect.com/science/article/pii/S0044848616303660?via%3Dihub
- Crosetti D. Biology, ecology and culture of grey mullet (Mugilidae). In: CRC Press Taylor & Francis Group; 2016. p. 42-127. Available from: https://books.google.co.in/books?hl=en&lr=&id=GAhCCwAAQBAJ&pg=PP1
- Koven W, Gisbert E, Meiri-Ashkenazi I, Nixon O, Israeli D, Tandler A, et al. The effect of weaning diet type on grey mullet (Mugil cephalus) juvenile performance during the trophic shift from carnivory to omnivory. Aquaculture. 2020;518:734848. doi:10.1016/j.aquaculture.2019.734848
- Boyd CE, D’Abramo LR, Glencross BD, Huyben DC, Juarez LM, Lockwood SG, et al. Achieving sustainable aquaculture: historical and current perspectives and future needs and challenges. J World Aquacult Soc. 2020;51:578-633. Available from: https://onlinelibrary.wiley.com/doi/10.1111/jwas.12714
- Alexander K, Potts T, Freeman S, Israel D, Johansen J, Kletou D, et al. The implications of aquaculture policy and regulation for the development of integrated multitrophic aquaculture in Europe. Aquaculture. 2015;443:16-23. Available from: http://dx.doi.org/10.1016/j.aquaculture.2015.03.005
- García-Márquez J, Cerezo IM, Figueroa LF, Abdala-Díaz RT, Arijo S. First evaluation of associated gut microbiota in wild thick-lipped grey mullets (Chelon labrosus, Risso 1827). Fishes. 2022;7:209. https://doi.org/10.3390/fishes7040209
- Cataudella S, Crosetti D, Massa F, editors. Mediterranean coastal lagoons: sustainable management and interactions among aquaculture, capture fisheries and the environment. Studies and Reviews; General Fisheries Commission for the Mediterranean. No. 95. Rome: FAO; 2015. p. 278. Available from: https://openknowledge.fao.org/server/api/core/bitstreams/2b9abb35-bb50-4c9d-bd7c-cbc98d21600c/content
- Caredda M, Addis M, Pes M, Fois N, Sanna G, Piredda G, et al. Physico-chemical, colorimetric, rheological parameters and chemometric discrimination of the origin of Mugil cephalus’ roes during the manufacturing process of Bottarga. Food Res Int. 2018;108:128-135. Available from: https://pubmed.ncbi.nlm.nih.gov/29735041/
- Khemis IB, Hamza N, Sadok S. Nutritional quality of the fresh and processed grey mullets (Mugilidae) products: a short review including data concerning fish from freshwater. Aquat Living Resour. 2019;32:2. Available from: https://www.alr-journal.org/articles/alr/abs/2019/01/alr180061/alr180061.html
- Vallainc D, Concu D, Gimenez G, Papiol GG, Loi B, Leggieri F, et al. Producing flat-head grey mullet Mugil cephalus (Linnaeus, 1758) fries in captivity from sexually mature adults collected in Sardinian lagoons. Aquac Rep. 2021;21:100844. Available from: https://www.sciencedirect.com/science/article/pii/S235251342100260X
- Pellizzato M. Classic and modern valliculture. In: The State of Italian Marine Fisheries and Aquaculture. Rome: Ministero delle Politiche Agricole, Alimentari e Forestali (MiPAAF); 2011; 237-238.
- Cataudella S, Tancioni L, Cannas A. Extensive aquaculture. In: Responsible Aquaculture towards Aquatic Production of the third Millennium. Rome: Unimar-Uniprom; 2001; 283-306. Available from: https://www.sidalc.net/search/Record/unfao:662493/Description
- Le MH, Wang D. Structure and membership of gut microbial communities in multiple fish cryptic species under potential migratory effects. Sci Rep. 2020;10:7547. Available from: https://www.nature.com/articles/s41598-020-64570-8
- Bi S, Yi H, Lai H, Li H, Liu X, Chen Q, et al. Intestinal microbiota of the four omnivorous fishes revealed by 16S rRNA metabarcoding from the habitats of oyster reefs. Ecol. Indic. 2023; 154: 110895. Available from: https://www.sciencedirect.com/science/article/pii/S1470160X23010373
- Cottiglia, M. Guides for the recognition of animal species in Italian lagoon and coastal waters - Lagoon fish. National Research Council. 1980;1:141. Available from: https://cir.nii.ac.jp/crid/1571417124807767296
- Trape S, Harrison IJ, Diouf PS, Durand JD. Redescription of Liza bandialensis (Teleostei: Mugilidae) with an identification key to mullet species of Eastern Central Atlantic. C R Biol. 2012;335(2):120-128. Available from: https://pubmed.ncbi.nlm.nih.gov/22325565/
- Hovda MB, Lunestad BT, Fontanilla R, Rosnes JT. Molecular characterisation of the intestinal microbiota of farmed Atlantic salmon (Salmo salar L.). Aquaculture. 2007;272(1-4):581-588. Available from: https://www.sciencedirect.com/science/article/abs/pii/S0044848607008927
- Skrodenyte-Arbaciauskiene V, Sruoga A, Butkauskas D, Skrupskeli K. Phylogenetic analysis of intestinal bacteria of freshwater salmon Salmo salar and sea trout Salmo trutta trutta and diet. Fish Sci. 2008; 74: 1307-1314. Available from: https://link.springer.com/article/10.1111/j.1444-2906.2008.01656.x
- Strickland JDH, Parsons TR. A Practical Handbook of Seawater Analysis, 2nd ed. Fisheries Research Board of Canada, Ottawa, ON, Canada. 1972;167. Available from: https://repository.oceanbestpractices.org/handle/11329/1994
- Martin M. Cutadapt removes adapter sequences form high-throughput sequencing reads. EMBnet.j. 2011;17:10-12. Available from: https://journal.embnet.org/index.php/embnetjournal/article/view/200
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Susan P Holmes, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. Available from: https://doi.org/10.1038/nmeth.3869
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:590–596. Available from: https://doi.org/10.1093/nar/gks1219
- McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013; 8: e61217. Available from: https://doi.org/10.1371/journal.pone.0061217
- Ofek T, Lalzar M, Laviad-Shitrit S, Izhaki I, Halpern M. Comparative Study of Intestinal Microbiota Composition of Six Edible Fish Species. Front Microbiol. 2021 Dec 7;12:760266. Available from: https://pubmed.ncbi.nlm.nih.gov/34950115/
- Floris R, Sanna G, Mura L, Fiori M, Culurgioni J, Diciotti R, Rizzo C, Lo Giudice A, Laganà P, Fois N. Isolation and Identification of Bacteria with Surface and Antibacterial Activity from the Gut of Mediterranean Grey Mullets. Microorganisms. 2021;9(12):2555. Available from: https://pubmed.ncbi.nlm.nih.gov/34946156/
- Bi S, Lai H, Guo D, Liu X, Wang G, Chen X, et al. The Characteristics of Intestinal Bacterial Community in Three Omnivorous Fishes and Their Interaction with Microbiota from Habitats. Microorganisms. 2021;9(10):2125. Available from: https://pubmed.ncbi.nlm.nih.gov/34683446/
- Kuang T, He A, Lin Y, Huang X, Liu L, Zhou L, et al. Comparative analysis of microbial communities associated with the gill, gut, and habitat of two filter-feeding fish, Aquaculture Reports 2020;18:100501. Available from: https://doi.org/10.1016/j.aqrep.2020.100501
- Katselis G, Koukou K, Dimitriou E, Koutsikopoulos C. Short-term seaward fish migration in the Messolonghi-Etoliko lagoons (Western Greek coast) in relation to climatic variables and the lunar cycle. Estuar Coast Shelf Sci. 2007;73:571-582. Available from: https://www.sciencedirect.com/science/article/abs/pii/S0272771407000649
- Ghanbari, M, Wolfgang K, Konrad J, Domig A. New view of the fish gut microbiome: Advances from next-generation sequencing. Aquaculture 2015;448:464-475. Available from: https://doi.org/10.1016/j.aquaculture.2015.06.033
- Egerton S, Culloty S, Whooley J, Stanton C, Ross RP. The Gut Microbiota of Marine Fish. Front Microbiol. 2018;9:873. Available from: https://pubmed.ncbi.nlm.nih.gov/29780377/
- Gadoin E, Durand L, Guillou A, Crochemore S, Bouvier T, Roque E, Dagorn L, Auguet JC, Adingra A, Desnues C, Bettarel Y. Does the Composition of the Gut Bacteriome Change during the Growth of Tuna? Microorganisms. 2021;9(6):1157. Available from: https://pubmed.ncbi.nlm.nih.gov/34072252/
- Speirs LBM, Rice DTF, Petrovski S, Seviour RJ. The Phylogeny, Biodiversity, and Ecology of the Chloroflexi in Activated Sludge. Front Microbiol. 2019;10:2015. Available from: https://pubmed.ncbi.nlm.nih.gov/31572309/
- Liang B, Wang LY, Zhou Z, Mbadinga SM, Zhou L, Liu JF, et al. High Frequency of Thermodesulfovibrio spp. and Anaerolineaceae in Association with Methanoculleus spp. in a Long-Term Incubation of n-Alkanes-Degrading Methanogenic Enrichment Culture. Front Microbiol. 2016;7:1431. Available from: https://pubmed.ncbi.nlm.nih.gov/27695441/
- Bovio P, Cabezas A, Etchebehere C. Preliminary analysis of Chloroflexi populations in full-scale UASB methanogenic reactors. J Appl Microbiol. 2019;126(2):667-683. Available from: https://pubmed.ncbi.nlm.nih.gov/30269410/
- Achenbach LA, Michaelidou U, Bruce RA, Fryman J, Coates JD. Dechloromonas agitata gen. nov., sp. nov. and Dechlorosoma suillum gen. nov., sp. nov., two novel environmentally dominant (per)chlorate-reducing bacteria and their phylogenetic position. Int J Syst Evol Microbiol. 2001;51(Pt 2):527-533. Available from: https://pubmed.ncbi.nlm.nih.gov/11321099/
- Dudley M, Salamone A, Nerenberg R. Kinetics of a chlorate-accumulating, perchlorate-reducing bacterium. Water Res. 2008;42(10-11):2403-10. Available from: https://pubmed.ncbi.nlm.nih.gov/18281075/
- Guarino F, Motta O, Turano M, Proto A, Vigliotta G. Preferential Use of the Perchlorate over the Nitrate in the Respiratory Processes Mediated by the Bacterium Azospira sp. OGA 24. Water 2020;12:2220. Available from: https://www.mdpi.com/2073-4441/12/8/2220
- Dasgupta PK, Martinelango PK, Jackson WA, Anderson TA, Tian K, Tock RW, Rajagopalan S. The origin of naturally occurring perchlorate: the role of atmospheric processes. Environ Sci Technol. 2005;39(6):1569-75. Available from: https://pubmed.ncbi.nlm.nih.gov/15819211/
- Kumarathilaka P, Oze C, Indraratne SP, Vithanage M. Perchlorate as an emerging contaminant in soil, water and food. Chemosphere. 2016;150:667-677. Available from: https://pubmed.ncbi.nlm.nih.gov/26868023/
- Gupta S, Lokesh J, Abdelhafiz Y, Siriyappagouder P, Pierre R, Sørensen M, Fernandes JMO, Kiron V. Macroalga-Derived Alginate Oligosaccharide Alters Intestinal Bacteria of Atlantic Salmon. Front Microbiol. 2019;10:2037. Available from: https://pubmed.ncbi.nlm.nih.gov/31572312/
- Liu H, Wang J, He T, Becker S, Zhang G, Li D, Ma X. Butyrate: A Double-Edged Sword for Health? Adv Nutr. 2018;9(1):21-29. Available from: https://pubmed.ncbi.nlm.nih.gov/29438462/
- Li X, Yan Q, Ringø E, Wu X, He Y, Yang D. The influence of weight and gender on intestinal bacterial community of wild largemouth bronze gudgeon (Coreius guichenoti, 1874). BMC Microbiol. 2016;16(1):191. Available from: https://pubmed.ncbi.nlm.nih.gov/27549138/
- Satish Kumar R, Kanmani P, Yuvaraj N, Paari KA, Pattukumar V, Arul V. Purification and characterization of enterocin MC13 produced by a potential aquaculture probiont Enterococcus faecium MC13 isolated from the gut of Mugil cephalus. Can J Microbiol. 2011;57(12):993-1001. Available from: https://pubmed.ncbi.nlm.nih.gov/22112158/
- Carnevali O, Maradonna F, Gioacchini G. Integrated control of fish metabolism, wellbeing and reproduction: The role of probiotic. Aquaculture 2017;472:144–155. Available from: https://doi.org/10.1016/j.aquaculture.2016.03.037

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.

 Save to Mendeley
Save to Mendeley
