Open Journal of Environmental Biology
Enrichment and remediation of uranium by microorganisms: A review
Qiuhan Yu1 and Ouyang Cui2*
2College of Mathematics and Statistics, Pingdingshan University, Pingdingshan 467000, PR China
Cite this as
Yu Q, Cui O (2023) Enrichment and remediation of uranium by microorganisms: A review. Open J Environ Biol 8(1): 020-038. DOI: 10.17352/ojeb.000037Copyright
© 2023 Yu Q, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.Uranium is a key raw material for the nuclear energy industry, the generation of nuclear energy is projected to double by 2040 to address the growing energy demands, which makes the uranium supply a matter of energy security. In addition, uranium is a heavy metal with both chemotoxicity and radiotoxicity, which seriously endangers human health and environmental safety. The growth in the utilization of uranium resources boosts the release of uranium into the environment. Therefore, the remediation of uranium contamination and recovery of uranium from a non-conventional approach is highly needed. Microorganism exhibits a high potential for immobilization of uranium. This review summarizes the ability of microorganisms to immobilize uranium from aqueous solutions and wastewater in terms of microbial species, performance, mechanism of enrichment and remediation, and applicable environment.
Introduction
Significance of uranium immobilization, utilization of uranium resources, remediation of environmental uranium pollution
Currently, nuclear energy is regarded as a relatively clean energy source due to the nature that is not affected by climate and geographic location, and increasingly valued by the national energy sector. Uranium, a natural radionuclide with a large energy density and a small carbon footprint, is the main raw material for generating nuclear energy, while the extremely low uranium concentration necessitates the development of a highly efficient strategy for uranium recovery.
What is more, human activities related to nuclear processes such as mining, fuel processing, weapons production, or nuclear accidents have caused uranium contamination of the environment [1]. Uranium is widely found in soil, rocks, oceans, and seas [2], and the most common isotopes of uranium in natural deposits are U-238, U-235, and U-234, and their proportions are 99.27%, 0.72%, and 0.0056%, respectively. Among these three isotopes, U-238 is considered to be stable with a half-life of 4.5 billion years [3]. It was reported that the average concentration of uranium (naturally occurring) in the earth's crust is 3 mg/kg [4]. In river water, the uranium concentration is between 0.01 and 6.6 μg/L, while under natural conditions, 30 and 3.32 μg/L are reported in groundwater and seawater, respectively [1]. The form of uranium mainly exhibited two oxidation states, U(IV) and U(VI) [1,5,6]. U(IV) is commonly found in ores in the form of uranium [UO2(s)] with insoluble and stable under anaerobic conditions. On the other hand, U(VI) is ubiquitous in aqueous systems under oxidizing conditions, and the morphology of this form is related to pH. Under acidic conditions (pH <5), U(VI) exists in a soluble form of UO22+ and is considered to be more toxic than U(IV) ions [7].
Uranium hydroxide with a pH between 5.0 and 7.0 is less toxic to aquatic plants and animals [1,8]. Uranium carbonate species, such as UO2(CO3)22and UO2(CO3)24-, dominate in rivers, ponds, lakes, and seawater above the environmental pH (> 7.5) [9].
Uranium is provided with radioactive and chemically toxic. Uranium-containing wastes released into the environment accumulate in soil or migrate to aquatic systems [2], and even may be introduced into terrestrial and aquatic food chains, posing a major threat to human, and environmental health and safety. Therefore, remediation of environmental uranium pollution is imminent [10,11]. In the process of uranium repair, the physicochemical processes are often cost-intensive. In addition, they were incapable of effectively removing a large number of toxic metals, resulting in the generation of by-products with danger. Problems associated with physicochemical processes can be overcome with the assistance of microbial systems [12]. Microorganisms interact with uranium to alter the concentration and/or oxidation state of the cell and its surrounding environment, thereby regulating the migration of uranium into the environment [13-16]. Despite the toxicity, local microorganisms exhibit high tolerance and survivability at sites contaminated with uranium. Various strategies and mechanisms have been adopted by microorganisms to persist in uranium-contaminated environments [17-19].
Microorganisms hold the ability to utilize their enzymatic reactions to precipitate, enrich, and redistribute uranium [20]. Therefore, the remediation technology of microbial has been considered as one of the most promising approaches. Various types of microorganisms have been reported to be involved in the uranium remediation process [21-23]. The bioreduction and biomineralization of uranium by microorganisms can relieve the toxic effect of uranium on microorganisms. Moreover, the active small molecule generated in the process of microbial metabolism can also bind to radionuclides and reduce the damage to microbial cells [24,25]. The bioprecipitation and reduction of uranium are the main forms of bioremediation of radionuclide uranium [26-28].
In addition to the bioremediation of the natural microbial, bioremediation of metal-contaminated environments using Genetically Engineered Microorganisms (GEMs) has become particularly important. Several reports emphasized the potential of GEMs in removing toxic metals and radionuclides [29-31]. Correlative genes have been expressed effectively in appropriate hosts, and the obtained GEMs showed excellent bioremediation efficiency [30,32-35]. Most studies concerning genetically engineered microorganisms focused on the precipitation of phosphatase-mediated [30,34-39]. Although genetically engineered microorganisms exhibited potential interactions with uranium, their development in bioremediation applications requires further validation.
Research status of uranium immobilization
Due to the importance of uranium in the nuclear power industry and the gradual reduction of uranium reserves in land ores, the unconventional strategies that efficiently recovery uranium are indispensable [40]. The ocean, containing 4.5 billion tons of uranium, possesses the largest uranium reserves on earth [41]. Recovering uranium from seawater was considered to be a potential strategy to meet the growing demand for uranium [42-44]. In addition, uranium as a heavy metal is extremely radioactive and chemically toxic. which brings serious damage to human health and the safety of the environment. Therefore, it is imminent to use cost-effective strategies to immobilize uranium from low-concentration uranium-containing environments or repair the environment contaminated by uranium.
Over the past 60 years, the immobilization of uranium and environmental remediation of uranium have received widespread attention [45,46]. A large number of adsorbents have been developed [47-50]. However, due to the cost-intensive of organic and inorganic adsorbents and the pollution of intermediates and by-products formed during the complex preparation process, therefore, these adsorbents are not suitable for large-scale application.
Due to its advantages of rapid growth, low cost, environmental friendliness, strong tolerance as well as the unique advantages in terms of the real-time, on-site treatment of large areas of contaminated water and soil, immobilization of uranium by biological entity seems to be a potential option. The properties of uranium adsorption by plants, algae, bacteria, fungi, and yeast have been reported several times [23,51-54]. They contain a variety of active groups for uranium adsorption and exhibit different mechanisms for uranium immobilization, such as bioreduction, bioprecipitation, and biosorption.
Benefits of microbial immobilization of uranium
In addition to the advantages of environmental friendliness, strong adaptability, and excellent tolerance mentioned earlier, immobilization of uranium by microbial still possesses other superiorities, such as small size (only a few micrometers), which can draw support from the functional groups on the cell surface, extracellular polymers and other substances to quickly adsorb and immobilize a certain amount of uranium in the solution, and then with the help of its unique metabolic effect to convert uranyl ions into a relatively stable state. What is more, microorganisms can be directly obtained through separation and culture, eliminating the need for other inorganic processing materials, so microorganisms were regarded as natural composite materials that exceeded the ability of chemists to construct materials.
Purpose of this review
Based on the environmental geochemical characteristics and pollution status of uranium in nature, this review summarizes the ability of microorganisms to immobilize uranium from aqueous solutions and seawater in terms of microbial species, performance, mechanism of enrichment, and applicable environment. Studying the interaction between uranium and microorganisms is of great significance for understanding the microbial environmental geochemical behavior of uranium, elucidating the laws of uranium migration and changes in the environment, and then developing remediation technologies that use microorganisms to regulate the migration and changes of uranium in the environment.
Performance of uranium immobilization by microorganisms
Applicable environment for uranium immobilization by microorganisms: Various external environments have great influences on the capacities of uranium immobilization of microorganisms. Krestou and Panias reported the calculated speciation of uranium as a function of concentration, pH, ionic strength, and carbonate [40]. In all instances, the free uranyl cation UO22+ is calculated at low pH <4.0. Hydrolysis of U(VI) increases and forms different hydroxyl complexes with the dominant species UO2(OH)5+ at pH 5.0, while UO2(CO3)34- is obtained above pH 8.5. However, the formation of polynuclear UO2CO3(OH)2- is enhanced over pH 6.0~8.0 as uranium concentration increased. It was reported that the optimal pH for uranium biosorption of microorganisms was ranging from 4.0 to 6.0 [53]. For example, the optimal pH for biosorption of uranium by Rhizopus arrhizus was between 4.0 and 5.0 [55], 5.6 to 6.0 for A. niger [56], and pH 5.0 for A. fumigatus [24] (Table 1). Salinity is another important factor governing uranium chemistry and concentration, thereby affecting adsorption performance. Several researches have reported that the presence of sodium chloride, calcium, magnesium, and bicarbonate significantly retards the uranium adsorption kinetics of adsorbents [57-61]. The influence of temperature on microorganism performance is truly multifaceted. Generally, the temperature of the medium is of great significance to the energy-dependent mechanism of microbial adsorption of metals [62-65]. Generally, due to the different mechanisms of uranium immobilization, the applicable environment for microbial immobilization of uranium differs in thousands of ways. Both biomineralization and accumulation require the formation of stable insoluble materials by reaction with phosphate to achieve the deposition of uranium and are greatly affected by pH [17,30]. For bioreduction, the oxidizing substances in the environment exhibited the most palpable effect on the immobilization of uranium [66]. In addition, due to the special environmental geochemical characteristics of U(VI), some coexisting ions also exhibit a certain effect on the reduction of uranium by microorganisms. Studies have found that Ca2+, Mg2+, CO32-, and NO3 possess a significant inhibitory effect on the reduction of uranium by microorganisms [67-69]. The first three mainly affected the reduction rate of uranium through the combination with uranium and under the condition of NO3- coexistence, NO3- replaced uranyl ions as the preferential electron acceptor in the process of microbial reduction of uranium, thereby affecting the reduction of uranium [58]. Except for the factors mentioned above, the activity or status of the cell posed a great effect on the performance of uranium biosorption, too. Generally, the dead cells exhibited a more excellent capacity than the alive cells [23,56,70-72].
Species of microorganisms with uranium immobilization activity: Biosorption is the property of living or dead biomass that absorbs and concentrates toxic metals from aqueous solutions [12,73-75]. In nature, there are a variety of biological materials that exhibit specific adsorption properties for uranium, such as bacteria [76,77], fungi [78,79], yeast [74,80,81], and algae [23,82]. In recent years, microorganisms have become an important alternative to remove radionuclides from aqueous solutions. Since no metabolic processes are involved, this phenomenon is usually rapid and growth-independent [75,83]. This section details the species of microorganisms with uranium immobilization activity.
Archaea and bacteria are ubiquitous and widespread in extreme cases [84,85]. They have a high surface-to-volume ratio and different functional groups (phosphate, carboxyl, amide, and hydroxyl) complexed with metals and radionuclides on the surface. Due to the lack of peptidoglycan in the cell walls of archaea, the cell wall characteristics of bacteria and archaea are different [86,87].
The performance of archaea in the biosorption of uranium has been confirmed recently. Two strains of Halobacterium noricense were capable of associating with uranium in a multistage process. After the initial phase of rapid adsorption, uranium was released, and then a slow recombination of uranium and cells was observed. In the first stage of biosorption, carboxylic acids and phosphorylated groups play an important role in uranyl binding [87].
Members of Bacillus, UUS-1, sp. dwc-2, and amyloliquefaciens have been proven to be effective uranium biosorbents and used for the biosorption of uranium several times [60,88,89]. The site-specific composite model initially described by scholars has been used to quantitatively predict the uranyl adsorption of Bacillus subtilis cells under different conditions. Ion strength had no effect on the adsorption performance, indicating that the inner-sphere surface complexation dominated the biosorption of U(VI) by Bacillus subtilis1 and the maximum adsorption capacity of B.subtilis at pH 4.5 and 298 K calculated by Langmuir model was 90.91 mg/g [90,91]. Recently, the role of immobilized B.subtilis alginate-chitosan microcapsules in uranium adsorption has been demonstrated and the protein surface layer (S layer) representing the outermost cell envelope component of bacteria is related to uranium biosorption [92]. In Bacillus JG-A12, the S layer binds uranium through phosphate and carboxyl groups [93,94]. The cells, spores, and S layer of this strain are embedded in silica gel, showing the potential of uranium adsorption from wastewater samples [93-95]. The strain UFO1 showed the potential to separate uranium under two different oxidation states, U(IV) and U(VI) [96]. Detailed analysis of uranium biosorption in Bacillus showed that it is not a single mode of binding but involved in a multiprocess of ion exchange, complexation, and bioaccumulation. 12 h was sufficient to achieve adsorption equilibrium and the uranium adsorption capacity was greater than 6.30 mg/g [97]. Bacillus mojavensis and Bacillus vallismortis loaded on multi-walled carbon nanotubes were used as solid-phase extractants to pre-concentrate uranium from aqueous solution and lake water samples [97-99].
Several actinomycetes have shown attractive potential for uranium adsorption [100]. For example, Streptomyces levophilus was demonstrated the basic characteristics of uranium adsorption from acidic solutions, which is mediated by the phosphate groups contained in the cell wall. These groups endow bacterial cells with a net negative charge and allow strong binding of positively charged uranyl ions in the pH range of 3.5 to 6.0 [100]. Similarly, the phosphodiester residues of the cell wall and cytoplasm fraction of Streptomyces longiformis bind uranium with significant efficiency [101]. Recent studies on Streptomyces spores dwc-3 have shown that at pH 3.0, uranium binds to amino, phosphate, and carboxyl groups in the cell wall, with the property of uranium adsorption greater than 3.0 mg/g [102]. It is reported that multifarious arthrobacter species were used for the biosorption of uranium and a large number of Arthrobacter were found in uranium-rich environments [7,103-105]. Arthrobacter ilicis accumulated uranium as precipitates within polyphosphate granules intracellularly [87]. The adsorption of uranium decreased in Arthrobacter G975 in the presence of an aqueous bicarbonate solution (a competitive ligand for U(VI)) [106]. The mobility of uranium is achieved in aquatic systems by forming highly soluble and stable uranyl carbonate complexes above neutral conditions, namely UO2CO3, UO2(CO3)22- and UO2(CO3)34- [107]. These uranyl species interfere with uranium complexation on the surface of microorganisms. Two more actinomycetes, Amycolatopsis sp. K47 and Brachybacterium sp. G1 recently has been proven to remove uranium by biosorption, too [87,108].
Two members of the Enterobacteriaceae family, Citrobacter and Serratia showed outstanding uranyl adsorption capacity [76,109]. U(VI) adsorption rate by Citrobacter freudii was found to be fast with carboxyl groups playing an important role in the combination of uranium [109].
The dead biomass of this bacterium showed better efficiency than living cells, indicating that uranium adsorption is not related to metabolic processes. A strain of Serratia marcescens showed high tolerance to uranium (4 mM) under acidic conditions (pH 3.5), removing 92% and 60-70% uranium from 100 µM and 2 mM uranyl solutions, respectively [76]. Similarly, a highly uranium-resistant strain Cuprius metallidurans CH34, resisting up to 30 mM uranium, immobilized U(VI) by complexing with phosphate radical or carboxyl groups of biomolecule containing lipopolysaccharide layers [77].
As for the member of Pseudomonas, the adsorption capacity of Pseudomonas putida to uranium has been confirmed [110-112]. Living or dead cells of P. putida are exposed to a mixed metal waste solution consisting of U, Pb, Cd, Zn, and Ni at pH 6.0 [112], showing high specificity for uranyl. Compared with the soil and clay components, dead cells of P. putida were more effective [113]. Pseudomonas aeruginosa CSU cells showed rapid intracellular accumulation of uranium within 10 s after exposure [80]. The same organism exhibits uranyl adsorption in the presence of transition metals under acidic and neutral conditions, which is equivalent to commercial cation exchange resins. However, certain cations [ferric iron (Fe3+) as a uranium analog] inhibited the binding of uranium to P. aeruginosa biomass in a significant manner [110]. Therefore, the removal of iron from wastewater is a prerequisite for the effective use of P. aeruginosa CSU biomass to adsorb uranium.
The uranium adsorption by Myxococcus xanthus depends on the age of the culture, uranyl ion concentration, and pH. Older cultures showed maximum adsorption when exposed to 1 mM uranium at pH 4.5. The adsorbed uranium is distributed on the cell wall and extracellular mucopolysaccharides [114]. Another strain of M. Xanthus performs uranium coordination with organic phosphate groups associated with the cell surface at a very low pH [115]. Most studies of uranyl binding of microorganisms are carried out in the pH range of 1.5 to 6.0, where uranyl ions and hydroxyl complexes are dominant [1,8]. The abilities of several microorganisms to sequester uranium above pH 7.0 were limited as a result of the repulsion between anionic carbonate complexes of uranium ([UO2(CO3)22-] and [UO2(CO3)34-]) and negatively charged cell surfaces [1,9,116]. However, two marine cyanobacteria have been proven to remove great quantities of uranium from aqueous solutions above pH 7.0 [43,84,117]. Synechococcus elongatus, when exposed to 100 μM uranium as a uranyl carbonate supplement at pH 7.8, showed rapid binding within 5 - 10 min [118]. The majority of bound uranium complexes with deprotonated carboxyl groups of amides and Extracellular Polysaccharides (EPS) can be released by HCl or EDTA [119]. Interestingly, it was observed that fixed Streptococcus elongatus cells were effective for uranium binding under continuous circulation conditions and could regenerated in multiple adsorption-desorption cycles (up to 3 times) without any significant loss of uranium binding above pH 7.0 [116]. Contrary to the rapid binding of uranium by Streptomyces elongatus, when the cells were exposed to 100 μM uranyl carbonate at pH 7.8 for 24 h, another marine cyanobacteria, Anabaena torulosa, showed that the accumulation of uranium in polyphosphates was especially slow [84,120].
Yeasts and filamentous fungi are lower eukaryotes that can be easily cultivated to produce large amounts of relatively inexpensive biomass that can effectively adsorb toxic metals and radionuclides. They are suitable for genetic manipulation and have a variety of biotechnology and industrial applications [74,121-123]. There are many reports on the biosorption of uranium by yeast and filamentous fungi in aqueous solution [12,22].
Some of the advantages of yeasts are cultivated easily on a large scale with highly productive, which makes them promising biosorbents for heavy metal immobilization [74]. The uranium adsorption properties of Saccharomyces cerevisiae have been reported several times [5,80,81,124]. Compared with living cells, dead cells of S. cerevisiae have a relatively higher metal biosorption capacity [125,126]. The heat-killed cells of S. cerevisiae release phosphate significantly due to membrane damage which complexed with uranyl residues and formed uranium phosphate nanoparticles on the cell surface [127]. The amidoxime group, known for its uranium selectivity, was successfully grafted onto S. cerevisiae and used for the adsorption of uranyl from salt lake brine solutions with excellent efficiency [128]. The dead biomass of S.cerevisiae with a rough surface and "nanopores" provides a larger contact area for uranyl adsorption than living cells with smooth surfaces [127]. Yarrowia lipolytica, a marine strain of the ascomycetous yeast, is currently being explored in the context of uranyl removal. Preliminary results demonstrated the ability of this strain to sequester 50% of the input uranium (50 μM uranium) loading up to 37.51 mg/g at pH 7.5 [129].
Lentinus sajor-caju is a saprophytic basidiomycetous white-rot fungus that easily grows on carbon sources and produces a variety of extracellular enzymes for the bioremediation of xenobiotics compounds [130]. Alkali-treated fungal biomass removed uranium more efficiently than untreated forms, with the superb efficiency of uranium adsorption of 268 mg/g at pH 4.5 [53]. The improved adsorption capacity of alkali-treated biomass is clearly the result of the increased availability of binding sites due to the deacetylation of chitin to chitosan. Schizophyllum commune, another basidiomycete fungus, showed that uranium accumulation intracellularly within the vacuoles and on the cell walls [131]. Phosphate groups instead of carboxyl groups promoted uranyl adsorption in S.comune, which is different from other uranyl microbial interactions, in which carboxyl groups played a major role in uranium complexation [118,132]. Rhizopus arrhizus was also provided with the nature of sequestering uranium from aqueous solutions and showed rapid adsorption for uranium, equilibrating within an hour with a maximum adsorption capacity of 180 mg/g at pH 4.0 [55,133]. Chitin and chitosan produced by fungi also exhibited the biosorption capacity for uranium [134-136].
Algae, which can be used as inexpensive biosorbent materials, are photosynthetic and autotrophic organisms generally distributed in aquatic environments. The biosorption properties of algae for uranium have been proposed.
Green algae is a kind of photosynthetic microorganism with various forms, from single-cell flagella to complex multicellular morphology. The living and dead cells of Chlorella vulgaris achieved rapid biosorption of uranium in a mineral medium with a low phosphate concentration of pH ranging from 3.0 to 6.0. Exposed to the uranium solution with the concentration of 0.1nM at pH 4.4, living cells bind 14.3mg/g dry biomass and dead cells 28.3 mg/g dry biomass within 5 min, which corresponds to 45% and 90% of total uranium in solution, respectively [23]. However, after a long period of cultivation, living cells caused the mobilization of bound uranium by releasing organic acids. C. vulgaris did not possess the capacity to adsorb uranium from natural or artificial seawater at pH 8.0. The adsorption of uranyl was inhibited due to the formation of stable uranyl-carbonate complexes [UO2(CO3)22-] and [UO2(CO3)34-] [137]. Brown algae are multicellular naturally and widely distributed in marine ecosystems [138]. Several brown algae have demonstrated the performance of uranium biosorption from aqueous solutions [139-142]. Under continuous flow conditions, the biomass of Sargassum fluitans was able to remove uranium up to 105 mg/g within 30 days [143]. Under acidic conditions of pH ranging from 2.5 to 4.0, Cystoseria indica exhibited excellent uranyl biosorption property of 233 mg/g at pH 4.0 [52,65,144,145]. The abundance of red algae in coastal and estuarine waters allows this biomass to be used as a cost-effective biosorbent. The biosorption of uranium by the red alga Catenella repens under acidic conditions has shown infusive results with a maximum loading of 303 mg/g from a solution of 100 mg/L uranium [146]. Another red algae, porphyridium cruentum, can achieve UO22+ ions uptake rapidly from ore and sludge [117].
Mechanisms of uranium immobilization: Microorganisms are complex in structure and exhibit various manners to combine with uranium under different conditions. At present, it is generally believed that the interaction between microorganisms and uranium can be divided into four mechanisms: bioreduction, cell surface adsorption, cell uptake and accumulation, and bioprecipitation [157,158]. Bioreduction, cellular uptake and accumulation, and bioprecipitation (biomineralization) can eventually lead to the deposition of uranium, and cell surface adsorption allows uranium to be adsorbed on the cell surface. In short, in the process of adsorption and immobilization, the role of microorganisms is mainly manifested in the following two ways:1) precipitation of uranium to prevent the migration and diffusion of uranium;2) rapid adsorption of uranium and timely recovery (Figure 1).
Microbial reduction of metals involves the immobilization of potentially toxic soluble metals and radionuclides in their insoluble form by alteration of their oxidation states. The microorganisms capable of reducing U(VI) are mainly anaerobic and facultative anaerobic bacteria [159] and also contain a small number of aerobic bacteria [160], archaea [87], and fungi [161]. The Extracellular Electron Transfers (EETs) mediated by C-type Cytochromes (C-Cyts) and microbial nanowires played a key role in the process of bacteria reducing U(VI). In recent years, the molecular biological mechanism involved in the U(VI) reduction process has been further explained.
Type of uranium reductive bacteria: In 1962, Woofolk, et al. determined the relationship between the amount of U(IV) produced in the crude extract of Veillonella alcalescens (formerly Micrococcus lactilyticus) and the amount of hydrogen consumption and confirmed that microorganisms can reduce U(VI) for the first time [162]. Subsequently, several studies proposed that anaerobic bacteria can use U(VI) as an electron acceptor to reduce U(VI) into a more stable U(IV) mineral. Research in the past 20 years has found that bacteria capable of reducing U(VI) mainly include but are not limited to Fe(III) reducing bacteria and sulfate-reducing bacteria. U(VI) reducing bacteria reported early are mainly distributed in the Proteobacteria and Firmicutes [163-166].
It has been found that anaerobic or facultative anaerobes can use U(VI) as the final electron acceptor and reduce it to pitch ore with the help of electronic shuttle media, such as cytochrome C, flagellum through enzymatic action [164,167-170]. Taking Shewanella as an example, the electrons generated by bacteria during metabolism are transferred along the respiratory chain, electrons are transferred to the coenzyme Q via NADH, and are released by the pigment Cym A on the plasma membrane of the cell to the periplasm. The transmembrane protein complex composed of three cytochrome molecules, Mtr A, Mtr B, and Mtr C, can transfer electrons from the periplasm to the cell surface. Omc A and Mtr C on the cell surface directly transfer electrons to U(VI) in the solution and reduce it to U(IV). Moreover, there is also a reduction of uranium in the periplasm when uranium enters the cell [171]. In addition, after microbial reduction, uranium formed non-pitch ore with phosphoric acid or carboxylic acid groups under the action of microorganisms [172]. Thermoterrabacterium ferrireducens, a thermophilic Gram-positive bacterium was reported to reduce insoluble (NH4) (UO2) (PO4)·3H2O to CaU4 (PO4)2·H2O, this was the first discovery that the final product of microbial reduction of uranium, in addition to UO2 previously considered by academics, and it also showed that the uranium phosphate minerals formed in uranium mining areas and uranium-contaminated areas may be further reduced [172].
Under acidic conditions ranging from pH 5.0 to 6.0, several species of Clostridia have demonstrated the inherent property of reducing U(VI) to U(IV) [17]. Clostridium is considered to be one of the most significant organisms to achieve uranium reduction under natural conditions [173-175], and the uranium reduction is related to Fermentative processes. In the absence of sulfate, Desulfotomaculum reducens MI-1 could grow with the help of U(VI) as the terminal electron acceptor [176]. This organism could couple the oxidation of organic compounds with the reduction of U(VI) to U(IV) and other metals manganese [Mn(IV) to Mn(II)], iron [Fe(III) to Fe(II)] or chromium [Cr(VI) to Cr(III)] for its growth. The spores of this bacterium could also give rise to the bioreduction of uranium by means of H2 as the electron donor [177]. Membrane-related electron transport respiratory (ETR) systems containing c-type cytochromes play a crucial role in uranium reduction [178]. Uranium-reducing facultative anaerobic bacteria prevent the formation of sulfides and hydrogen sulfide complexes, which have toxic and inhibitory effects on cellular metabolism [179]. In such microorganisms, extracellular U(VI) reduction was related to the flow of electrons through NADH-dehydrogenase, which is a primary electron donor associated with the ETR system. Salmonella subterranean isolated from nitrate and uranium-contaminated subsurface sediments is an acid-tolerant bacterium and an important component of U(VI) reducing enrichment culture, showing notable properties of uranium reduction at pH 4.5 [180]. In the bioreduction of uranium, members of the genus Shewanella have been comprehensively studied. Preliminary studies on Shewanella putrefaciens indicated that the bioreduction of uranium was promoted by enzymatic reactions coupled with electron transport chains in the organism [166]. Shewanella alga is an iron-reducing bacterium that can lead to the reduction of uranium when complexed with multidentate aliphatic ligands (malonate, oxalate, and citrate) rather than monodentate acetate groups [181], however, it was incapable of reducing uranium complex with aromatic ligand and metal chelator. Another vital element that affects the bioreduction kinetics of uranium is the bioavailability of uranium to the bacteria. In the presence of sodium bicarbonate (NaHCO3), the kinetics of U(VI) reduction by Shewanella oneidensis MR-1 became slow. At pH 7.0, with the increase of NaHCO3 concentration, the amount of uranium adsorbed decreases due to the formation of negatively charged soluble uranyl carbonate species [182].
A range of biological and chemical redox transformations were performed in S.oneidensis, including subsequent reduction to U(IV) nanoparticles [uraninite (UO2)], absorption of U(VI) to the cell surface, and formation of U(VI) nanowires [metaschoepite (UO3.2H2O)], providing a novel strategy for the bioremediation of uranium in contaminated aquifers [183]. The two-step uranium reduction was superior to the one-step reduction process that was previously reported [163]. Mixed bacterial cultures isolated from natural sediments and pure cultures of Desulfosporosinus species, Geobacter sulfurreducens, S.oneidensis MR-1, and S.putrefaciens CN32 are also reported to possess the property of forming biogenic uraninite nanoparticles [184-187]. In S.oneidensis MR-1, c-type cytochromes played a vital role in reducing U(VI) to extracellular UO2 nanoparticles [188]. Compared with the wild-type strains, the reduction of uranium of mutants of S. oneidensis MR-1 which lacks omcA or mtrC showed slower [188]. Similarly, the endecaheme c-type cytochrome present in the OM of Shewanella sp. participated in the extracellular reduction of iron [Fe(III)] oxides and uranium [U(VI)] [189]. With the assistance of outer membrane c-type cytochromes present in the extracellular polymeric substances, biofilms of Shewanella sp. HRCR-1 was also capable of reducing uranium efficiently [190]. Nevertheless, S. oneidensis MR-1 and its biofilm displayed boundedness in U(VI) reduction in flow and batch reactors [191]. Studies have shown that in addition to genetic manipulations, alternative organic materials/modules can also promote and accelerate the rates of uranium bioreduction.
Anthraquinone-2,6-disulfonate as a humus substitute enhanced the rates of uranium reduction in S. oneidensis [18]. Similarly, flavin mononucleotide secreted by Shewanella species improved the reduction rate of U(VI) [192].
Except for the Gram-positive bacteria above-mentioned, Gram-negative bacteria Geobacter metallireducens, Anaeromyxobacter dehalogenans as well as various sulfate-reducing bacteria (SRB) also displayed great potential in uranium bioreduction. Geobacter species can mediate the coupled reduction of Fe(III) and U(VI) under anaerobic subsurface conditions. G. metallireducens, a Fe (III) reducing bacterium, exhibited a direct reduction of U(VI) instead of an indirect reduction of Fe(III). The reduction process was conducive to the growth of bacterial and the culture ceased to grow once uranium was depleted from the growth medium [163]. This organism reduced uranium in groundwater samples and the deposits of uraninite were observed on cell surfaces [166]. Nitrate, a co-contaminant associated with uranium, is detrimental to the reduction processes and it was essential to add sufficient amounts of acetate (as an electron donor) initially to reduce nitrate [193]. Reduction and precipitation of uranium on surfaces rich in electron donors bring about detriment to the cell envelope and interfere with the associated functions. For the sake of safeguarding cellular integrity and viability, conductive pili in Geobacter sp. have been reported as the performance of extracellular reduction of U(VI) to U(IV) [194]. Pili were considered to be the cardinal uranium reductases that accepted electrons from the cell envelope and c-type cytochromes. These acted as electrical conduits between the cells and uranium. The loss of pili (Pili A-mutant) caused damage to the reduction of uranium in Geobacter and resulted in the enhancement of periplasmic mineralization, which, in turn, reduced cell viability and cellular respiration [195]. G. sulfurreducens, a sulfate-reducing species of Geobacter, initially reduced U(VI) to a U(V) intermediate which resulted in the formation of tetravalent uranium on disproportionation [184]. Cytochromes associated with outer surfaces (c-type) played an important role in uranium reduction in this organism [159]. Moreover, periplasmic cytochrome MacA, a diheme c-type cytochrome peroxidase, was found to be essential for U(VI) reduction in G. sulfurreducens, and a fraction of the reduced uranium was localized in the periplasm. Growth yields of Geobacter lovleyi and G. sulfurreducens were decreased when U(VI) was used as an electron acceptor, indicating that this process of reduction imposed an additional burden on the growing cells [196]. The chemical state of biomass and enrichment mechanism of U(VI) during bioconcentration was analyzed by XPS and the results showed that uranium displayed two valence states (U(VI) and U(IV)). After bioconcentration, the resistant Absidia corymbifera showed the reductive effect of U(VI) to U(IV), which is also a resistance mechanism for the reduction of uranium hazards by Absidia corymbifera [197]. Certain studies have shown that U(VI) in groundwater is removed after in situ prompting metal reduction, which is usually accompanied by a significant increase in the growth and activity of different metal-reducing microorganisms in Deobacteraceae. The analysis of XPS and FTIR results showed that the removal of U(VI) by the Absidia corymbifera may be complicated [198].
Under microcosm and in situ conditions, the role of Geobacter species in uranium bioremediation was essential. Microcosm studies showed the enrichment of the members of the Geobacteraceae family could reduce uranium under low bicarbonate concentrations (1 mM) in the presence of ethanol as the electron donor [199]. With the supplementation of acetate as an electron donor, concentrations of U(VI) were observed to decrease from 0.4 to those below 0.18 μM which was believed to be the maximum contaminant limit [200].
An anaerobic myxobacterium isolated from Oak Ridge FRC, TN, U.S. was identified as A. dehalogenans by using H2 as the electron donor to reduce uranium rather than acetate which was utilized by Geobacter [201]. Some electron acceptors, such as Fe(III) citrate or Fe(III) oxide, exhibited an inhibitory effect on the reduction of U(VI) in A. dehalogenans [201]. In the presence of lactate or H2 acted as electron donors, the SRB and Desulfovibrio desulfuricans have been reported to achieve the reduction of uranium rapidly and directly. U(VI) and sulfate reduction occurred simultaneously and reduced uranium was observed in the form of extracellular uraninite. D.desulfuricans cells contained within a semipermeable membrane showed a rapid reduction of uranium at concentrations as high as 24 mM [165]. Bicarbonate extracted uranium from contaminated soils and subsequently microbial reduced with D. desulfuricans showing a potential strategy for enriching uranium from contaminated soil and sediments [202]. Cytochrome c3 seemed to be a component of the electron pathway in vivo that participated in the reduction of U(VI) to U(IV) in D. desulfuricans. In the case of lactic or pyruvate as the electron donor, the cytochrome c3 mutant of D. desulfuricans G20 is not as effective in reducing uranium as the wild-type strain (almost 50%) [66]. The capacities of uranium reduction of various sulfur-reducing bacteria were analyzed [164]. Among different isolates, Desulfovibrio vulgaris was the most effective uranium-reducing organism followed by Desulfovibrio baculatus, Desulfovibrio sulfodismutans, and Desulfovibrio baarsii [164]. Uranium reduction by all these SRBs relied on the supply of electron donors D.vulgaris was incapable of growing in the case of U(VI) as the sole electron acceptor, precipitated uranium as uraninite and both sulfate and uranium reduced simultaneously which was similar to D.desulfuricans [164]. Another SRB, Desulfosporosinus sp. and its type strain Desulfosporosinus orientis showed the capacity for enzymatic reduction of uranium in the presence of lactate or H2 as an electron donor [7].
In the presence of acetate, glucose, or ethanol, a field study associated with the growth and activity of indigenous microorganisms was performed [203]. The replenishment of these electron donors brought out the development of an anaerobic and reducing environment that was conducive to the reduction of NO3-, Fe(III), and U(VI). NO3- dependent, microbial-mediated U(IV) oxidation is an important process in regulating the stability of U(VI) reduction in places contaminated with high concentrations of nitrates. Supplementation with acetate, and in situ uranium bioremediation trials were carried out. Rapid reduction of uranium prevalent in the form of Ca-UO2-CO3 ternary complexes was achieved by iron and uranyl respiring Geobacter species [204]. In subsequent studies, at the same location, in the presence of acetate, lactate, hydrogen release compound (HRC), or vegetable oil, uranium reduction was observed [205]. Another report described U(VI) bioreduction over an extended period. Emulsified vegetable oil amendments caused effective bioreduction at depths up to 50 m [206]. Recent studies have confirmed U(VI) bioreduction in iron oxide-rich sediments following ethanol supplementation [207]. The major problem associated with in situ bioremediation of uranium-contaminated sites is the fate of U(IV) induction in sub-surfaces which re-oxidizes to U(VI) form and affects the process economy negatively.
Uranium bio precipitation refers to the direct entry and immobilization of U(VI) into mineral lattice under the action of microorganisms. Unlike the bioreduction of uranium, U(VI) does not accept electrons in this process. Most of the uranium bio precipitation is aerobic and facultative aerobic microorganisms. The process and mechanism of interaction between U(VI) and microorganisms were studied and found that the uranium bio-precipitation process is involved in the microbial adsorption of U(VI), coordination of functional groups with uranium, and stress regulation of phosphatases.
The number of aerobic microorganisms exhibited an excellent adsorption capacity for uranium. Chen, et al. found that Bacillus thuringiensis could reach a high uranium removal rate within 2 h with a maximum capacity of 416 mg/g biomass. The functional groups present on the cell surface play an important role in the microbial immobilization of U(VI). In the initial stage of interaction between U(VI) and microorganisms, U(VI) is first quickly adsorbed on the surface of microorganisms by electrostatic action, and then further immobilized by functional groups such as peptidoglycan, amino group, carboxyl group, and phosphate group on the cell surface [161]. Choudhary and Sar found that Pseudomonas aeruginosa isolated from uranium slag contains carboxyl, amino, and phosphoric acid groups, which are the dominant functional groups for U(VI) immobilization. Two types of Bacillus sphaericus JG-7B and Sphingomonas sp. S15-S1 which was isolated from extreme environments exhibited the capacity of U(VI) precipitation on the surface or inside of cells. Potentiometric titration analysis shows that the organic functional groups on the cell surface (carboxyl, amino, hydroxyl, and phosphate groups) are the primary binding sites of uranium [208]. Screening of the functional groups directly on the cell surface weakens the ability of microorganisms to adsorb heavy metals to a certain extent [209].
In addition to the functional groups, phosphatase also plays an important role in the bioprecipitation of uranium. The overexpression of phosphatase as a result of heavy metal stimulation could be used as a detoxification mechanism for bacteria. Phosphate produced by intracellular phosphatase under the stimulation of U(VI) promoted the immobilization of uranium better, which can reduce the toxicity of U(VI) to cells to some extent. About 30 years ago, Macaskie, et al. discovered that Citrobacter sp. could achieve the transformation of U(VI) into U(VI) phosphate minerals under the mediation of phosphatase. Subsequently, it was reported that phosphatase could cooperate with extracellular lipopolysaccharides to promote the conversion of U(VI) to NH4UO2PO4 [19]. In addition, phosphatase was also involved in the immobilization of U(VI) to phosphate in Bacillus sphaericus JG-7B and Sphingomonas sp. S15-S1 [208]. Islam, et al. studied the effects of uranium on a variety of bacteria isolated from uranium deposits and found that most of them could immobilize uranium into uranyl phosphate compounds or uranyl phosphate saline compounds in a phosphorus-free solution, so it was speculated that these bacteria might induce uranyl ion immobilization by increasing the concentration of surrounding phosphate under the action of phosphatase [210]. A study on the sorption mechanism of Kocuria toward U(VI) indicated that uranium adsorption by –P = O, -OH, -C = O, -COOH at a short time and then forming CaU(PO4)2 precipitation in the surface. Thus, the effect of sorption by Kocuria cells not only depends on the passive adsorption of active sites but also on the release of phosphate from the cell. Fast immobilization of U(VI) on the cell surface firstly and gradually more phosphate was released by Kocuria and was immobilized with U(VI) as the species of uranium phosphate with very low solubility [148]. The process of uranium mineralization was investigated by Saccharomyces cerevisiae and the toxicity experiments showed that the viability of the cell was not significantly affected by 100 mg/L U(VI) under 4 days of exposure time. Moreover, the batch experiments showed that the phosphate concentration and pH value rose over time during U(VI) adsorption [54]. Meanwhile, thermodynamic calculations demonstrated that the adsorption system for UO2HPO4 was supersaturated. The X-ray Diffraction (XRD), Fourier Transform Infrared (FTIR), Field Emission Scanning electron Microscope (FE-SEM) equipped with Energy Dispersive X-ray spectroscopy (EDX), and X-ray Photoelectron Spectroscopy (XPS) analysis indicated that the U(VI) was first attached onto the cell surface and reacted with the groups that exist on cell surface, such as hydroxyl, carboxyl, and phosphate groups, through electrostatic interactions and complexation. As the immobilization of U(VI) transformed it from an ionic to an amorphous state, lamellar uranium precipitate came into being on the cell surface. With the prolongation of time, the amorphous uranium compound vanished, and some crystalline substances were observed extracellularly, which were well-identified as tetragonal-chernikovite. Furthermore, the size of chernikovite was regulated by cells at the nano-scale, ultimately, the perfect crystal was generated [156]. Radiation-resistant Deinococcus and E. coli could immobilize uranium in the form of uranium phosphate and the phosphatases in both bacteria. In addition to bacteria, the interaction and mineralization between fungi represented by yeasts and uranium have also attracted the attention of some scholars. A study on the interaction between a variety of selected yeasts and uranium mineralization found that yeasts could utilize phosphatase to decompose phosphates produced by organic phosphorus sources, thereby immobilizing uranium on the cell surface in the form of phosphate. A yeast stimulated by uranium can release a large amount of phosphate under the action of phosphatase. These generated phosphates will be transferred from the cell to the extracellular and bound to uranium. After 4 days of action, hydrogen uranium mica [H2(UO2)2(PO4)2·8H2O] with obvious crystal will be formed outside the cell [211]. The research mentioned above indicated that phosphatase immobilized uranium mainly through phosphate produced by the decomposition of organophosphorus sources, and the addition of glycerol phosphate and other substances as the organophosphorus source in the process of immobilization of uranium by aerobic bacteria could facilitate the immobilization of uranium by bacteria better [212]. In oxygen-containing and low pH environments, the low solubility of U(VI)-phosphate minerals makes it a good form of U(VI) immobilization [213]. There is increasing research on the interaction between aerobic bacteria and uranium, but the mechanism of aerobic bacteria immobilizing uranium still has a lot of research space. Li, et al. found that the accumulation of uranium by Streptomyces was related to the surface functional groups and may also involved in the ion exchange process [102]. The first study by Bader, et al. found that the interaction between archaea and uranium was a multi-stage process, which may include adsorption under the action of functional groups and mineralization after adsorption, but the specifics of these two processes are yet to be studied [214]. In another study, Gerber, et al. found that the interaction between Acidovorax facilis and uranium consisted of an adsorption and post-adsorption equilibrium process [215]. Transmission electron microscopy analysis showed that uranium-containing substances accumulated outside and inside the cell, which was related to phosphorus, but the specific mechanism of action still needed further exploration. Moreover, the previous research on the biomineralization of uranium mostly focused on the observation of the cell surface. Some studies have found that the product of bacterial immobilization of uranium existed in the cell, but there is no detailed study and explanation of how these substances are produced [208]. Pan, et al. conducted electron microscopy and XRD analysis on the interaction of two other strains of Bacillus thuringiensis with U(VI) and found that in the initial stage of the interaction between bacteria and uranium, uranium was first adsorbed on the cell surface in the form of an amorphous substance. This amorphous material gradually entered the cell and converted into NH4(UO2) (PO4)·3H2O minerals stored in the cell around 10 nm [216]. This process of intracellular and extracellular transformation suggested that aerobic bacteria may not only be able to immobilize uranium but also could immobilize nano-scale uranium minerals in micro-scale cells so that they could be stored better, which was important for reducing the mobility of uranium. However, there was no further study on how the uranium-containing material transferred from extracellular to intracellular.
Whether the phenomenon of uranium immobilization in cells, intracellular and extracellular transfers was a special case or universal and what role enzymes play in them required further research. For some uranium-contaminated soil and water that were not suitable for bioreduction, bioprecipitation provided novel strategies, but its related mechanism had not been fully clarified. The functional groups and phosphatases contained in microorganisms played a significant role in the immobilization of uranium, and the formation of uranium-phosphate products is a relatively common phenomenon. To a certain extent, relative to microbial reduction, whether it is in the size and stability of the uranium product or the possible storage form of the product, there is a relatively large exploration space for uranium bio-precipitation.
By summarizing the research results of the four mechanisms in recent years, it is found that the treatment of uranium-containing wastewater by microorganisms was not the role of a single mechanism, but a joint action of multiple mechanisms. Microorganisms could adsorb uranium on the surface of cells through surface adsorption; When the phosphorus source supplementation was sufficient in the organism, the immobilization of uranium could be more and more stable; Under reducing conditions, microorganisms could survive through the biological reduction process, simutaneously, completing the immobilization of uranium; Biomineralization could act as a supplementary technology for bioreduction, and the concentration of uranium ions can be maximized regardless of the reduction or oxidation conditions.
Microbial reaction to uranium immobilization
As we all know, uranium can cause liver, lung, and kidney damage in humans [10,11]. Although many microorganisms have been proven to respire uranium, the toxicity of uranium in microorganisms has been confirmed from the inhibition of microbial activity, cell surface deformation and loss of cell viability, transcription and translation process suspension, growth arrest, DNA replication suspension, and oxidative damage [74,106]. For the purpose of resisting toxicity and reducing the harm caused by the presence of uranium, microorganisms have undertaken a series of stress responses to counteract, showing the potential of uranium immobilization and bioremediation.
Growth change and the responses of physiological and biochemical: Transcriptomics studies were conducted on Metallosphaera prunae exposed to high concentrations of soluble uranium (1,238 mg/L) and found that within 15 min of uranium exposure, large amounts of cellular RNA degradation and the termination of transcription and translation processes in the organism, which may be to resist uranium toxicity [219]. It was pointed out that siderophores may play a vital role in the uranium chelation of M. prunae because it was found that genes related to the iron complex transport system were significantly induced during uranium exposure [219]. It was reported that when exposed to U(VI), the growth rate of G.sulfreducens was slightly reduced, and numerous proteins that related to central metabolism were in low abundance. Moreover, phosphoenolpyruvate synthase, a protein that is involved in translation, ribosome biogenesis, and amino acid biosynthesis were all in less abundance after exposure to uranium [220,221]. The morphological changes of B.amyloliquefaciens rich in U(VI) were determined using scanning electron microscopy and energy dispersive spectroscopy, the results showed that the morphological characteristics of B. amyloliquefaciens after bioconcentration were significantly different from those before, in which the cell surface is smooth and rod-like. However, after only 3 h of exposure to 200 mg/L of uranium, the bacterial surface became rougher and damaged fragments of cells existed on the bacterial surface. Moreover, in the absence of uranium, the surface of B.amyloliquefaciens exhibited a clear cell wall structure, and some organelles inside the cell were also localized, the morphology of the whole cell was complete. Under 200 mg/L of uranium stress, part of the uranium ion enters the cytoplasm first, As the inner organelle and cell wall of the cell are destroyed, the whole cell interior becomes a homogeneous system, losing the protection function, thus making it easier for uranium to enter the cell interior and combine with the intracellular desmin, making the cell lose its function, and eventually leading to cell disintegration [60]. The same phenomenon occurs in C.Utilis, under the action of uranium stress, intracellular biomolecules of C.Utilis combine with U(VI) and even lead to the breaking of biochemical bonds of biomolecules, organelle disintegration, impaired cell function, resulting in metabolic effects, and ultimately hindering the growth of C.Utilis. In addition, the effects of different concentrations of uranium on the growth of C.Utilis were diverse. The biomass of C.Utilis, MDA (Malondialdehyde), H2O2, GSH (Glutathione), and SOD were measured under different concentrations of uranium. Changes in the concentrations or contents of GSH and SOD with the increase of U(VI) concentration, the microbial biomass gradually decreased, and the contents of MDA and H2O2 increased first and then decreased, indicating that the ability of active oxygen cleared of C.Utilis cells under U(VI) stress is time-delayed. GSH and SOD reflect the ability of cells to remove reactive oxygen species, and the changes of the two have greater synchronization [222]. A number of previous studies have found that many microorganisms are destroyed to varying degrees following heavy metal or radionuclide enrichment, such as Arsenic enrichment in R.oryzae, Cobalt enrichment in A.niger, and U(VI) enrichment in Mucor [58,223]. Another study on the immobilization of uranium by Bacillus velezensis Strain UUS-1 showed that under the stress of uranium with the concentration of 10 mg/L, the morphology of bacterial cells was changed and more flagella was observed, which plays a vital role in the movement of the bacterium to a suitable environment for growth. As a heavy metal, a high concentration of uranium was possibly toxic to biological entities, and UUS-1 generated more flagella to escape the high uranium environment is undoubtedly a self-protect mechanism [88]. A study on the bioaccumulation and transformation of U(VI) by sporangiospores of Mucor circinelloides showed that the intracellular and extracellular morphological structure changed significantly, and levels of intracellular H2O2, O3-, GPx, and SOD compounds in sporangiospores increased significantly under U(VI) stress [149]. What is more, the resistance of M.circinelloides to U(VI) and As(V) stress is mainly attributed to the biosynthesis of thiol compounds (NP-SH and PBSH), activation of antioxidant enzymes (SOD and CAT), and secretion of organic acids (oxalic acid and citric acid) [58].
In summary, the radionuclide U(VI) has the ability to inhibit the growth of cells and induce the production of reactive oxygen species.
Differential expression of key biomolecules: It is possible to understand the expression of genes/proteins under different stress conditions with the development of analytical and bioinformatics techniques. The identification of functional genes and proteins implicated in metal responses is fundamental in deciphering underlying molecular mechanisms and for developing in situ bioremediation strategies.
For the purpose of assessing how bacterial cells respond to the presence of U(VI), G.sulfurreducens that were grown anaerobically to the mid-exponential phase were exposed to 100 mM uranyl acetate for 4 h and then collected for proteomic analysis. A total of 1363 proteins were detected in cells. There were 203 proteins detected with higher abundance during exposure to U(VI) compared with the control cells and 148 proteins with lower abundance. This accounted for 26% of the total proteins detected, indicating that protein expression was significantly affected by the presence of U(VI). Proteins associated with energy conservation had the highest number of proteins with greater abundance following uranium exposure. However, in the presence of U(VI), the proteins involved in translation, and ribosome biogenesis were all in less abundance, which reflected that protein biosynthesis in the presence of U(VI) also seems to be less important [224].
Although there are no known uranium-specific detoxification systems in microorganisms, metal efflux pumps for other toxic metals exist and could conceivably play a role in preventing uranium toxicity. Several efflux pumps in the RND (resistance-nodulation-cell division) family, which confer metal tolerance by extruding a wide spectrum of metals, were more abundant in cells exposed to U(VI) [224,225]. Many other proteins related to the binding and transport of metals were also significantly more abundant in the presence of U(VI), such as the putative periplasmic tungstate ABC transporter, which is part of the tungstate transport complex, and MgtA commonly involved in Mg2+ transport [224]. Besides, G.sulfreducens showed increased levels of several efflux pumps belonging to the RND family of superoxide dismutase and superoxide reductase upon exposure to uranium [226]. Proteins involved in phosphate and iron metabolism were found to be abundant in a uranium-tolerant bacterium. Microbacterium oleivorans A9 and the role of siderophores were proposed in uranium transportation in this bacterium [227]. In addition, many proteins related to the regulation of gene expression in response to changes in the environment were differentially expressed when cells were exposed to U(VI). For instance, the transcriptional regulator of the Fur family was expressed in higher abundance when U(VI) was present. The fur regulon is of great significance in the regulation of the iron uptake pathway, indicating iron uptake increase under U(VI) exposure [228]. Two transcriptional regulatory proteins of the G.sulfurreducens TetR family that coordinate the expression of efflux pumps were also expressed in higher abundance. The TetR family has a hand in the regulation of efflux pumps and tolerance to toxic compounds [229]. The transcriptional regulators of the ArsR family, which responds to metal ion stress and modulates the transcription of genes involved in metal efflux, sequestration, and detoxification, were expressed in high abundance [230-233].
For the detoxification of heavy metal, another strategy is precipitation [171,234]. It is understood that several microorganisms were able to use the phosphate derived from polyphosphates to precipitate uranium [13]. In the cells that were exposed to uranium, polyphosphate kinase and exopolyphosphatase, which catalyze the transfer of phosphate from ATP to the formation of long-chain polyphosphates and irreversibly hydrolyzes polyP to form phosphate, respectively, both exhibited higher abundance, indicating its excellent prospect in uranium detoxification. In addition, polyphosphates can indirectly affect the outflow of other heavy metals, such as cadmium in E. coli, Anacystis nidulans, and nickel in Staphylococcus aureus, thereby regenerating ATP, which can then be used to activate the outflow of ATPase [235-237]. Proteome analysis of C. crescentus exposed to uranium revealed the down regulation of cell cycle regulators, motility, chemotactic proteins, and up regulation of possible phytase [238]. Phytase provides the necessary phosphate groups for uranium precipitation and facilitates the survival of C. crescentus cells in the presence of uranium [239]. The raise of protein expression involved in general stress response or reactive oxygen species detoxification was observed in uranium-exposed cells of A. ferrooxidans [240]. Similarly, E. coli cells exposed to uranyl ions showed different accumulation of oxidative stress proteins and other proteins such as NADH/quinone oxidoreductase WrbA [241].
Uranium has a high affinity for organic molecules and is capable of forming strong bonds with functional groups in proteins [242]. The uranium binding can bring out changes in the conformation of proteins [243,244]. Uranium ions can produce ligands containing functional groups of thiolates as well as carboxylate from acidic amino acids [243,245,246]. Enzymes in G. sulfurreducens that contribute to protein folding may help to avoid these potential pernicious effects. For example, exposure to U(VI) resulted in higher expression of the chaperonin GroES, the DnaJ-related molecular chaperone, and the DnaJ adenine nucleotide exchange factor that is involved in the protection and renaturation of heat-labile proteins [224]. This is in consistent with a former study which reported that transcripts of DnaJ and GrpE were found to be expressed in higher abundance in cells of the dissimilatory metal-reducing bacterium Shewanella oneidensis strain MR-1 [182]. The expression of several proteins related to peptide secretion and trafficking was also more abundant in the presence of U(VI). For example, SecE and SecF, which belong to the general Sec system, and PulQ and GspK, which are part of the type II secretion system, were in higher abundance in the presence of U(VI) [224]. Previous studies have suggested that the type II secretion system has an essential role in localizing several metal-containing proteins on the outer surface of the cell [247,248]. Besides protein, uranium also possesses a high affinity for DNA, which can result in DNA strand breakage and inhibition of DNA-protein interactions [242,244,245,249].
Exposing G. sulfurreducens to U(VI) resulted in a higher abundance of both superoxide dismutase (sodA) and superoxide reductase [226]. A former transcriptional study of the Geobacter species reported that the gene encoding the superoxide dismutase was highly expressed despite the presence of a highly reduced environment [250]. Another study evaluating the transcriptional expression of the Geobacter uraniireducens also showed that the sodA gene was upregulated when the isolate was grown in the contaminated subsurface sediments [251]. Both results suggested that the expression of the superoxide dismutase could not only be triggered by oxygen stress but also by other factors in the sediments. Furthermore, the gene encoding the superoxide dismutase upregulated when cells of the highly uranium-tolerant oligotrophy, Caulobacter crescentus, were exposed to uranium, cadmium, chromate, and dichromate, suggesting that this enzyme is involved in the response to a wide range of heavy metals [252].
Perspective and outlook
Microbes possess many advantages in treating low-concentration uranium-containing wastewater. However, it is only in the stage of experience and laboratory, and there are still many challenges and critical issues in practical and industrial applications that need to be addressed further. It can be studied from the following aspects:
- The particle size of the microbial reduction and non-reduction products of uranium is relatively small, and it is easy to migrate or oxidize in the environment. How to regulate the process of crystallization and whether the particle size of the crystal can be changed by artificial regulation becomes a concern.
- Nowadays, the research on microbial immobilization of uranium is mostly focused on the role of anaerobic bacteria, while immobilization of uranium by aerobic is relatively few. But, in the actual environment, there are many aerobic bacteria in the surface and shallow water, and the treatment of uranium pollution by anaerobic bacteria alone may not meet the needs of surface treatment. However, in the process of immobilizing uranium, aerobic bacteria often sacrifice their own phosphate, resulting in their survival rate decreased. It may be possible to consider artificial regulation to improve the survival rate of bacteria while better-assisting bacteria to immobilize uranium. Some studies have observed the migration and transformation of uranium in and out of cells during the process of microbial mineralization, but there is no explanation about how this process occurs and what the influencing factors are.
- Single microorganisms and the microbial community stability. The immobilization mechanism of different types of microorganisms on uranium still needs to be studied further. The type and quantity of ligands on the cell surface affected the surface adsorption capacity, while the specific influence means is not yet clear. It is possible with the help of modern instrument analysis to analyze and modify the groups on the cell surface that interact with uranium to obtain better adsorption materials. Using genetic recombination technology to cultivate "super strains" with high adsorption performance and low environmental pollution can be more effective in treating low-concentration uranium-containing wastewater.
- Microbial treatment of uranium is usually by a variety of mechanisms, how to combine bioreduction and biomineralization together to achieve a better ability to remove uranium is one of the research directions in the future. In addition, it is found that the microbial-mediated uranium mineralization process is the result of a variety of microorganisms performing different mechanisms. Therefore, it is not sufficient to study uranium immobilization by a single microorganism. Studies are carried out from this perspective and based on this to explore the diversity of microorganisms to enhance the stability of the microbial system used to treat uranium-containing wastewater, thereby enhancing the ability of uranium immobilization.
Conclusion
The microbial adsorption of uranium is a complex process, and its adsorption mechanism is not singular in most cases. Only from the perspective of the molecular to study biological adsorption mechanism, the chemical composition and biological synthesis pathway of the effective adsorption of the cell wall can find out the microbial and its environment that is suitable for adsorption of uranium, thereby improving the adsorption selectivity of adsorbents on the specific ion. Various testing and analysis techniques can be used to study the binding sites and forms of uranium on the surface and interior of cells, the changes in binding energy between uranium and specific functional groups in cells, as well as the structure and characteristics of functional groups. With the continuous development of genetic engineering technology, strengthening genetic engineering research, creating gene libraries, and constructing engineering bacteria with strong adsorption capacity are the future development directions.
The author would like to acknowledge the financial support of Pingdingshan university.
Data availability statement
The manuscript is a review article so data sharing is not applicable to this article. No new data were created or analyzed in this manuscript.
Notes on contributors
Yu Qiuhan is working as a lecturer at Pingdingshan University. She has a PhD Degree in Biomaterials. She has worked in the area of water/wastewater treatment, uranium extraction, and biomaterials development for five years.
- Markich SJ. Uranium Speciation and Bioavailability in AquaticSystems: An Overview. Sci. World J. 2002; 2:707-729.
- Gavrilescu M, Pavel LV, Cretescu I. Characterization and Remediation of Soils Contaminated With Uranium. J. Hazard. Mater. 2009; 163:475-510.
- Wong S. Introductory nuclear physics. John Wiley & Sons. 1987.
- Kalin M, Wheeler WN, Meinrath G. The removal of uranium from mining waste water using algal/microbial biomass. J. Environ. Radioact. 2005; 78 (2): 151-177.
- Lu X, Zhou XJ, Wang TS. Mechanism of uranium(Ⅵ) uptake by Saccharomyces cerevisiae under environmentally relevant conditions:Batch, HRTEM, and FTIR studies. J. Hazard. Mater. 2013; 262:297-303.
- Bernhard G, Geipel G, Brendler V, Nitsche H. Uranium speciation in waters of different uranium mining areas. J. Alloys Compd. 1998; 271(10):201-205.
- Suzuki Y, Banfield JF. Resistance to, and Accumulation of, Uranium by Bacteria from a Uranium-Contaminated Site. Geomicrobiol. 2004; 21(2):113-121.
- Choppin GR. Actinide speciation in the environment. Journal of Radioanalytical & Nuclear Chemistry. 2007; 273(3):695-703.
- Konstantinou M, Pashalidis I. Speciation and spectrophotometric determination of uranium in seawater. Cartographic Journal the World of Mapping. 2004; 5(1):217-228.
- Anke M, Seeber O, Müller R. Uranium transfers in the food chain from soil to plants, animals, and man. Chemie Der Erde Geochemistry. 2009; 69 (1):75-90.
- Priest ND. Toxicity of depleted uranium. Lancet. 2001 Jan 27;357(9252):244-6. doi: 10.1016/s0140-6736(00)03605-9. PMID: 11214120.
- Wang J, Chen C. Biosorption of heavy metals by Saccharomyces cerevisiae: A review. Biotechnol. Adv. 2006; 24(5):427-451.
- Merroun ML, Selenska-Pobell S. Bacterial Interactions with Uranium: An Environmental Perspective. J. Contam. Hydrol. 2008; 102(3-4):285-295.
- Majumder E, Wall JD. Uranium Bio-Transformations: Chemical or Biological Processes? J. Inorg. Chem. 2017; 7(2):28-60.
- Macaskie L, Empson R, Cheetham A. Uranium bioaccumulation by a Citrobacter sp. as a result of enzymatically mediated growth of polycrystalline HUO2PO4. Science. 1992; 257: (5071);782-784.
- Acharya C. Uranium Bioremediation: Approaches and Challenges. Springer International Publishing. 2015.
- Gao W, Francis AJ. Reduction of Uranium(VI) to Uranium(IV) by Clostridia. Appl. Environ. Microbiol. 2008; 74 (14):4580-4584.
- Liu JX, Xie SB, Wang YH. U(VI) reduction by Shewanella oneidensis mediated by anthraquinone-2-sulfonate. Transactions of Nonferrous Metals Society of China. 2015; 25(12):4144-4150.
- Km ML, Yong P, Goddard DT. Enzymatically mediated Bio-precipitation of uranium by a Citrobacter sp.: a concerted role for exocellular lipopolysaccharide and associated phosphatase in biomineral formation. Microbiol. 2000; 146 (8):1855-1867.
- Nevin KP, Finneran KT, Lovley DR. Microorganisms Associated with Uranium Bioremediation in a High-Salinity Subsurface Sediment. Appl. Environ. Microbiol. 2003; 69(6):3672-3675.
- Taillefert M, Keaton M. Cappellen; Philippe; DiChristina; Thomas, Effects of aqueous uranyl speciation on the kinetics of microbial uranium reduction. Geochim. Cosmochim. Acta. 2015.
- Kapoor A, Viraraghavan T. Fungal biosorption -an alternative treatment option for heavy metal bearing wastewaters: a review. Bioresour. Technol. 1995; 53 (3):195-206.
- Vogel M, Günther A, Rossberg A. Biosorption of U(VI) by the green algae Chlorella vulgaris in dependence on pH value and cell activity. Sci. Total Environ. 2010; 409(2):384-395.
- Bhainsa KC, Souza SF. Biosorption of uranium(VI) by Aspergillus fumigatus. Biotechnol. Tech. 1999; 13(10):695-699.
- Mumtaz S, Streten JC, Parry DL. Fungi outcompete bacteria under increased uranium concentration in culture media. J. Environ. Radioact. 2013; 120C:39-44.
- Cao B, Ahmed B, Kennedy DW. Contribution of Extracellular Polymeric Substances from Shewanella sp. HRCR-1 Biofilms to U(VI) Immobilization. Environmental Science & Technology. 2011; 45(13):5483-5490.
- Krawczyk BE, Grossmann K, Arnold T. Influence of uranium (VI) on the metabolic activity of stable multispecies biofilms studied oxygen microsensors and fluorescence microscopy. Geochi.Cosmochi.Acta. 2008; 72(21):0-5265.
- Behrends T, Arnold T. Implementation of microbial processes in the performance assessment of spent nuclear fuel repositories. Appl. Geochem. 2012; 27(2):453-462.
- Garbisu C, Alkorta I. Utilization of genetically engineered microorganisms (GEMs) for bioremediation. J. Chem. Technol. Biotechnol. 1999; 74(7) :599-606.
- Kulkarni S, Ballal A, Apte SK. Bioprecipitation of uranium from alkaline waste solutions using recombinant Deinococcusradiodurans. J. Hazard. Mater. 2013; 262:853-861.
- Meltem UD, Benjamin S. Use of Genetically Engineered Microorganisms (GEMs) for the Bioremediation of Contaminants: Critical Reviews in Biotechnology. Crit. Rev. Biotechnol. 2006; 26(3).
- Nilgiriwala KS, Anuradha A, Amara SR. Cloning and overexpression of alkaline phosphatase PhoK from Sphingomonas sp. strain BSAR-1 for bio-precipitation of uranium from alkaline solutions. Appl. Environ. Microbiol. 2008; 74(17):5516-23.
- Kulkarni S, Misra CS, Gupta A. Interaction of uranium with bacterial cell surface: Inferences from phosphatase mediated uranium precipitation. Appl. Environ. Microbiol. 2016; 82(16):4965-4974.
- Appukuttan D, Rao AS, Apte SK. Engineering of Deinococcus radiodurans R1 for Bioprecipitation of Uranium from Dilute Nuclear Waste. Applied & Environmental Microbiology. 2007;73(4):1393-1393.
- Appukuttan D, Seetharam C, Padma N. PhoN-expressing, lyophilized, recombinant Deinococcus radiodurans cells for uranium bioprecipitation. J. Biotechnol. 2011; 154(4):285-290.
- Basnakova G, Stephens ER, Thaller MC. The use of Escherichia coli bearing a phoN gene for the removal of uranium and nickel from aqueous flows. Appl. Microbiol. Biotechnol. 1998; 50(2):266-272.
- Battista JR. Against all odds: The survival strategies of Deinococcus radiodurans. Annual Review of Microbiology. 1997.
- Misra CS, Mukhopadhyaya R, Apte SK. Harnessing a radiation-inducible promoter of Deinococcus radiodurans for enhanced precipitation of uranium. J. Biotechnol. 2014; 189: 88-93.
- Barak Y, Ackerley DF, Dodge CJ. Analysis of Novel Soluble Chromate and Uranyl Reductases and Generation of an Improved Enzyme by Directed Evolution. Appl. Environ. Microbiol. 2006; 72(11):7074-7082.
- Klaus M, Maria W. Uranium from German Nuclear Power Projects of the 1940s- A Nuclear Forensic Investigation. Angew. Chem. Int. Ed. 2015; 54(45):13452-13456.
- Lindner H, Schneider E. Review of cost estimates for uranium recovery from seawater. Energy Economics. 2015; 49:9-22.
- Sholl DS, Lively RP. Seven chemical separations to change the world. Nature. 2016; 532(7600):435-437.
- Tabushi I, Kobuke Y, Nishiya T. Extraction of uranium from seawater by polymer-bound macrocyclic hexaketone. Nature. 1979; 280(5724): 665-666.
- Parker BF, Zhang Z, Rao LF. An overview and recent progress in the chemistry of uranium extraction from seawater. Dalton Trans. 2018; 47(3):639-644.
- Abney CW, Mayes RT, Saito T. Materials for the Recovery of Uranium from Seawater. Chem. Rev. 2017; 117(23):13935-14013.
- Yue Y, Mayes RT, Kim J. Seawater Uranium Sorbents: Preparation from a Mesoporous Copolymer Initiator by Atom-Transfer Radical Polymerization. Angew. Chem. Int. Ed. 2013; 52(50):13458-13462.
- Yuan YH, Niu BY, Yu, QH. Photoinduced Multiple Effects to Enhance Uranium Extraction from Natural Seawater by Black Phosphorus Nanosheets. Angew. Chem. Int. Ed. 2020; 132(3).
- Yuan YH, Yu QH, Wen J. Ultrafast and Highly Selective Uranium Extraction from Seawater by Hydrogel-like Spidroin-based Protein Fiber. Angew. Chem. Int. Ed. 2019; 58(34).
- Ma CX, Gao JX, Wang D. Sunlight Polymerization of Poly(amidoxime) Hydrogel Membrane for Enhanced Uranium Extraction from Seawater. Adv. Sci. 2019; 6(13).
- Chen L, Bai Z, Zhu L. Ultrafast and Efficient Extraction of Uranium from Seawater Using an Amidoxime AppendedMetal–Organic Framework. ACS Appl. Mater. Interfaces. 2017; 9(38):32446-32451.
- Li X, Liu Y, Ming ZH. Adsorption of U(VI) from aqueous solution by cross-linked rice straw. J. Radioanal. Nucl. Chem. 2013;298(1):383-392.
- GoeK C, Aytas S, Sezer H. Modeling uranium biosorption by Cystoseira sp. and application studies. Sep. Sci. 2017;52(5):792-803.
- Bayramo LG, Elik GK, Arica MY. Studies on accumulation of uranium by fungus Lentinus sajor-caju. J. Hazard. Mater. 2006; 136(2):345-353.
- Ohnuki T, Ozaki T, Yoshida T. Mechanisms of uranium mineralization by the yeast Saccharomyces cerevisiae. Geochim. Cosmochim. Acta. 2005; 69(22):0-5316.
- Tsezos M, Volesky B. The Mechanism of Uranium Biosorption by Rhizopus arrhizus. Biotechnol. Bioeng. 1982; 24(2):385-401.
- Ding H, Zhang X, Yang H. Biosorption of U(VI) by active and inactive Aspergillus niger: equilibrium, kinetic, thermodynamic, and mechanistic analyses. J. Radioanal. Nucl. Chem. 2019; 319(3):1261-1275.
- Liu L, Zhang Z, Song W. Removal of radionuclide U(VI) from aqueous solution by the resistant fungus Absidia corymbifera. J. Radioanal. Nucl. Chem. 2018; 318(2).
- Song W, Wang X, Chen Z. Enhanced immobilization of U(VI) on Mucor circinelloides in the presence of As(V): Batch and XAFS investigation. Environmental Pollution. 2018; 237(6):228-236.
- Yan B, Sun Q, Wang S. Characterization of nano-iron oxyhydroxides and their application in UO22+ removal from aqueous solutions. J. Radioanal. Nucl. Chem. 2011.
- Liu L, Liu J, Liu X. Kinetic and equilibrium of U(VI) biosorption onto the resistant bacterium Bacillus amyloliquefaciens. J. Environ. Radioact. 2019; 203:117-124.
- Yao T, Wu X, Chen X. Biosorption of Eu(III) and U(VI) on Bacillus subtilis: Macroscopic and modeling investigation. J. Mol. Liq. 2016; 219:32-38.
- Bayramo LG, Elik GK, Arica MY. Studies on accumulation of uranium by fungus Lentinus sajor-caju. J. Hazar. Mater. 2006; 136(2):345-353.
- Tsezos M, Volesky B. Biosorption of uranium and thorium. 1981.
- Arica MY, Bayramoglu G, Yilmaz M. Biosorption of Hg2+, Cd2+, and Zn2+ by Ca-alginate and immobilized wood-rotting fungus Funalia trogii. J. Hazard. Mater. 2004; 109(1-3):191-199.
- Khani MH, Keshtkar AR, Ghannadi M. Equilibrium, kinetic, and thermodynamic study of the biosorption of uranium onto Cystoseria indica algae. J. Hazard. Mater. 2008; 150(3): 612-618.
- Rayford B, Gentry DM. Uranium Reduction by Desulfovibrio desulfuricans Strains G20 and a Cytochrome c[sub 3] Mutant. Appl. Environ. Microbiol. 2002.
- Finneran KT, Housewright ME, Lovley DR. Multiple influences of nitrate on uranium solubility durin~1 bioremediation of uranium-contaminated subsurface sediments. Environ. Microbiol. 2002; 4(9):510-516.
- Brooks SC, Fredrickson JK, Carroll SL. Inhibition of Bacterial U(VI) Reduction by Calcium. Environ.Sci. Technol. 2003; 37(9):1850-1858.
- Istok JD, Senko JM, Krumholz LR. In Situ Bioreduction of Technetium and Uranium in a Nitrate-Contaminated Aquifer. Environ.Sci. Technol. 2004; 38(2):468-475.
- Castro GI, Rojas VG, Quintero ZI. A Comparative Study on Removal Efficiency of Cr(VI) in Aqueous Solution by Fusarium sp and Myrothecium sp. Water Air Soil Pollut. 2017; 228(8).
- Cephidian A, Makhdoumi A, Mashreghi M. Removal of anthropogenic lead pollution by a potent Bacillus species AS2 isolated from the geogenic contaminated site. Int. J. Environ. Sci. Technol. 2016; 13(9):2135-2142.
- Song W, Wang X, Sun Y. Bioaccumulation and Transformation of U(VI) by Sporangiospores of Mucor circinelloides. Chem. Eng. J. 2019; 362(8).
- White C, Gadd GM. Biosorption of Radionuclides by Fungal Biomass. J. Chem. Technol. Biotechnol. 2007; 49(4):331-343.
- Volesky B, Holan ZR. Biosorption of Heavy Metals. Biotechnol. Progr. 1995; 11(3):235-250.
- Lloyd JR, Macaskie LE. Chapter 11 Biochemical basis of microbe-radionuclide interactions. Elsevier Science & Technology: 2002.
- Kumar R, Acharya C, Joshi SR. Isolation and analyses of uranium tolerant Serratia marcescens strains and their utilization for aerobic uranium U(VI) bioadsorption. J. Microbiol. 2011; 49(4):568-574.
- Llorens I, Untereiner G, Jaillard D. Uranium Interaction with Two Multi-Resistant Environmental Bacteria: Cupriavidus metallidurans CH34 and Rhodopseudomonas palustris. Plos One. 2012; 7(12):e51783.
- Rodr GE, Nuero O, Guillén F. Degradation of phenolic and non-phenolic aromatic pollutants by four Pleurotus species: the role of laccase and versatile peroxidase. Soil Biology & Biochemistry 2004; 36(6):909-916.
- Akhtar K, Akhtar MW, Khalid AM. Removal and recovery of uranium from aqueous solutions by Trichoderma harzianum. Water Res. 2007; 41(6):1366-1378.
- Strandberg GW, Shumate SE, Parrott JR. Microbial cells as biosorbents for heavy metals: accumulation of uranium by Saccharomyces cerevisiae and Pseudomonas aeruginosa. Appl. Environ. Microbiol. 1981; 41(9).
- Liu M, Dong F, Yan X. Biosorption of uranium by Saccharomyces cerevisiae and surface interactions under culture conditions. Bioresour.Technol. 2010; 101(22):8573-8580.
- Aytas S, Gunduz E, Gok C. Biosorption of Uranium Ions by Marine Macroalga Padina pavonia. Clean Soil Air Water. 2014; 42(4): 498-506.
- Ahluwalia SS, Goyal D. Microbial and plant-derived biomass for removal of heavy metals from wastewater. Bioresour. Technol. 2007; 98(12):2243-2257.
- Acharya C, Chandwadkar P, Apte SK. Interaction of uranium with a filamentous, heterocystous, nitrogen-fixing cyanobacterium, Anabaena torulosa. Bioresour.Technol. 2012; 116:290-294.
- Jroundi F, Merroun ML, Arias JM. Spectroscopic and Microscopic Characterization of Uranium Biomineralization in Myxococcus xanthus.
- Vieira RH, Volesky B. Biosorption: A solution to pollution? International Microbiology the Official Journal of the Spanish Society for Microbiology. 2000; 3(1).
- Bader M, Müller K, Foerstendorf H. Comparative analysis of uranium bioassociation with halophilic bacteria and archaea. Plos One. 2018; 13(1):e0190953.
- Yuan YH, Yu QH, Yang S. Ultrafast Recovery of Uranium from Seawater by Bacillus velezensis Strain UUS-1 with Innate Antibiofouling Activity. Adv. Sci. 2019; 6(18).
- Li X, Ding C, Liao J. Biosorption of uranium on Bacillus sp. dwc-2: a preliminary investigation on mechanism. J. Environ. Radioact. 2014; 135:6-12.
- Gorman LD, Fein JB, Soderholm L. Experimental study of neptunyl adsorption onto Bacillus subtilis. Geochim. Cosmochim. Acta. 2005; 69(20):4837-4844.
- Gorman LD, Elias PE, Fein JB. Adsorption of Aqueous Uranyl Complexes onto Bacillus subtilis Cells. Environ. Sci. Technol. 2005; 39(13):4906-4912.
- Ke T. Preparation and biosorption evaluation of Bacillus subtilis alginate-chitosan microcapsule. Nanotechnol.Sci.Appl. 2017; 10:35-43.
- Raff J, Soltmann U, Matys S. Bacteria-based bioremediation of uranium mining wastewaters by using sol-gel ceramics. Springer Berlin Heidelberg. 2002.
- Merroun ML, Raff J, Rossberg A. Complexation of Uranium by Cells and S-Layer Sheets of Bacillus sphaericus JG-A12. Appl. Environ. Microbiol. 2005; 71(9):5532-5543.
- Raff J, Soltmann U, Matys S. Biosorption of Uranium and Copper by Biocers. Chem. Mater. 2003; 15(1):240-244.
- Ray AE, Bargar JR, Sivaswamy V. Evidence for multiple modes of uranium immobilization by an anaerobic bacterium. Geochim. Cosmochim. Acta. 2011; 75(10):0-2695.
- Ozdemir S,Oduncu MK, Lilinc E. Resistance, bioaccumulation, and solid phase extraction of uranium (VI) by Bacillus vallismortis and its UV-vis spectrophotometric determination. J. Environ. Radioact. 2017; 171:217-225.
- Sadin, Özdemir MK, Ersin K. Tolerance and bioaccumulation of U(VI) by Bacillus mojavensis and its solid phase preconcentration by Bacillus mojavensis immobilized multiwalled carbon nanotube. J. Environ. Manage. 2017; 187:490-496.
- Kilinc E. GeoBacillus thermoleovorans immobilized on Amberlite XAD-4 resin as a biosorbent for solid phase extraction of uranium (VI) prior to its spectrophotometric determination. Microchimica Acta. 2018; 178(3-4):389-397.
- Tsuruta T. Adsorption of uranium from acidic solution by microbes and effect of thorium on uranium adsorption by Streptomyces levoris. J. Biosci. Bioeng. 2004; 97(4):275-277.
- Friis N, Myers KP. Biosorption of uranium and lead by Streptomyces longwoodensis. Biotechnol. Bioeng. 1986;28(1):21-28.
- Li X, Ding C, Liao J. Bioaccumulation characterization of uranium by a novel Streptomyces sporoverrucosus dwc-3. J. Environ.Sci. 2016; 41(3):162-171.
- Kumar R, Nongkhlaw M, Acharya C. Uranium (U)-Tolerant Bacterial Diversity from U Ore Deposit of Domiasiat in North-East India and Its Prospective Utilisation in Bioremediation. Microbes.Environ. 2013; 28(1):33-41.
- Miller CL, Landa ER, Updegraff DM. Ecological aspects of microorganisms inhabiting uranium mill tailings. Microb. Ecol. 1987; 14(2):141-155.
- Chapon V, Piette L, Vesvres MH. Microbial diversity in contaminated soils along the T22 trench of the Chernobyl experimental platform. Appl. Geochem. 2012; 27(7):1375-1383.
- Carvajal DA, Katsenovich YP, Lagos LE. The effects of aqueous bicarbonate and calcium ions on uranium biosorption by Arthrobacter G975 strain. Chem. Geol. 2012; 330:51-59.
- Langmuir D. Uranium solution-mineral equilibria at low temperatures with applications to sedimentary ore deposits. Geochim. Cosmochim. Acta. 1978; 42(6):547-569.
- Celik F, Camas M, Kyeremeh K. Microbial Sorption of Uranium Using Amycolatopsis sp. K47 Isolated from Uranium Deposits. Water Air Soil Pollut. 2018; 229(4).
- Xie S, Yang J, Chen C. Study on biosorption kinetics and thermodynamics of uranium by Citrobacter freudii. J. Environ. Radioact. 2008; 99(1):126-133.
- Hu MZ, Norman JM, Faison BD. Biosorption of uranium by Pseudomonas aeruginosa strain CSU: Characterization and comparison studies. Biotechnol. Bioeng. 1996; 51(2):237-247.
- Choi J, Park JW. Competitive adsorption of heavy metals and uranium on soil constituents and microorganisms. Geosci. J. 2005; 9(1):53-61.
- Choi J, Ju YL, Yang JS. Biosorption Of Heavy Metals And Uranium By Starfish And Pseudomonas Putida. J. Hazard. Mater. 2009; 161(1):157-162.
- Choi J, Park JW. Competitive adsorption of heavy metals and uranium on soil constituents and microorganisms. Geosciences Journal. 2005; 9(1):53-61.
- González-Muñoz MT, Merroun ML, Omar NB. Biosorption of uranium by Myxococcus xanthus. Int. Biodeterior. Biodegrad. 1997; 40(2):107-114.
- Jroundi F, Merroun ML, Arias JM. Spectroscopic and Microscopic Characterization of Uranium Biomineralization in Myxococcus xanthus. Geomicrobiol. J. 2007; 24(5):441-449.
- Acharya C, Apte SK. Insights into the interactions of cyanobacteria with uranium. Photosynth. Res. 2013;118(1-2):83-94.
- Acharya C, Chandwadkar P, Joseph D. Uranium (VI) recovery from saline environment by a marine unicellular cyanobacterium, Synechococcus elongatus. J. Radioanal. Nucl. Chem. 2012; 295(2):845-850.
- Acharya C, Joseph D, Apte SK. Uranium sequestration by a marine cyanobacterium, Synechococcus elongatus strain BDU/75042. Bioresour. Technol. 2009; 100(7):2176-2181.
- Acharya C, Chandwadkar P, Joseph D. Uranium (VI) recovery from saline environment by a marine unicellular cyanobacterium, Synechococcus elongatus. Journal of Radioanalytical & Nuclear Chemistry. 2013; 295(2):845-850.
- Acharya C, Apte SK. Novel surface-associated polyphosphate bodies sequester uranium in the filamentous, marine cyanobacterium, Anabaena torulosa. Metallomics. 2013; 5(12):1595-1598.
- Bai RS, Abraham TE. Studies on chromium(VI) adsorption–desorption using immobilized fungal biomass. Bioresour.Technol. 2003; 87(1):17-26.
- Zinjarde SS. Food-related applications of Yarrowia lipolytica. Food Chem. 2014; 152:1-10.
- Zinjarde S, Apte M, Mohite P. Yarrowia lipolytica and pollutants: Interactions and applications. Biotechnol. Adv. 2014; 32(5):920-933.
- Faghihian H, Peyvandi S. Adsorption isotherm for uranyl biosorption by Saccharomyces cerevisiae biomass. J. Radioanal. Nucl. Chem. 2012; 293(2):463-468.
- Soares EV, Coninck GD, Duarte F. Use of Saccharomyces cerevisiae for Cu2+ removal from solution: the advantages of using a flocculent strain. Biotechnol. Lett. 2002; 24(8):663-666.
- Xia L, Tan K, Wang X. Uranium Removal from Aqueous Solution by Banyan Leaves: Equilibrium, Thermodynamic, Kinetic, and Mechanism Studies. J. Environ. Eng. 2013; 139(6):887-895.
- Wang T, Zheng X, Wang X. Different biosorption mechanisms of Uranium(VI) by live and heat-killed Saccharomyces cerevisiae under environmentally relevant conditions. J.Environ.Radioact. 2017; 167:92-99.
- Bai J, Yin X, Zhu Y. Selective uranium sorption from salt lake brines by amidoximated Saccharomyces cerevisiae. Chem. Eng. J. 2016; 283:889-895.
- Zinjarde SS, Pant AA. Hydrocarbon degraders from tropical marine environments. Mar. Pollut. Bull. 2002; 44(2):118-121.
- Rodr GE, Nuero O, Guillén F. Degradation of phenolic and non-phenolic aromatic pollutants by four Pleurotus species: the role of laccase and versatile peroxidase. Soil Biol. Biochem. 2004; 36(6):909-916.
- Alix G, Johannes R, Mohamed L. The interaction of U(VI) with the Schizophyllum commune was studied by microscopic and spectroscopic methods. BioMetals. 2014; 27(4):775-785.
- Merroun M, Hennig C, Rossberg A. Characterization of U(VI)-AcidithioBacillus ferrooxidans complexes using EXAFS, transmission electron microscopy, and energy-dispersive X-ray analysis. Radiochim. Acta. 2009; 91(10):583-592.
- Tsczos M. The role of chitin in uranium adsorption by R. arrhizus. Biotechnol. Bioeng. 1983; 25(8):2025-2040.
- Tsezos M, Georgousis Z, Remoudaki E. Mechanism of aluminum interference on uranium biosorption by Rhizopus arrhizus. Biotechnol. Bioeng. 1997; 55(1):16-27.
- Takashi S, Takao H, Akira N. Adsorption of Uranium by Chitin Phosphate and Chitosan Phosphate. Agricultural & Biological Chemistry. 2014.
- Wang G, Liu J, Wang X. Adsorption of uranium (VI) from aqueous solution onto cross-linked chitosan. J. Hazard. Mater. 2009; 168(2-3):1053-1058.
- Nakajima A, Korikoshi T, Sakaguchi T. Uptake of Manganese Ion by Chorella regularis. Agricultural & Biological Chemistry. 1979.
- Rousvoal S, Groisillier A, Dittami SM. Mannitol-1-phosphate dehydrogenase activity in Ectocarpus siliculosus, a key role in mannitol synthesis in brown algae. Planta. 2011; 233(2):261-273.
- Khani M. Statistical analysis and isotherm study of uranium biosorption by Padinasp. algae biomass. Environ. Sci. Pollut. Res. 2011; 18(5):790-799.
- Eiif G, Sule A. Biosorption of Uranium Ions by Marine Macroalga Padina pavonia. Clean Soil Air Water A Journal of Sustainability & Environmental Safety. 2014.
- Gok C, Aytas S. Biosorption of uranium(VI) from aqueous solution using calcium alginate beads. J. Hazard. Mater. 2009; 168(1):369-375.
- Yu J, Wang J, Jiang Y. Removal of Uranium from Aqueous Solution by Alginate Beads. Nucl. Engineer. Technol. 2016; S1738573316301826.
- Yang J, Volesky B. Modeling Uranium-Proton Ion Exchange in Biosorption. J. Environ. Sci. Technol. 1999; 33(22):4079-4085.
- Ghasemi M, Keshtkar AR, Dabbagh R. Biosorption of uranium(Ⅵ) from aqueous solutions by Ca-pretreated Cystoseira indica alga: Breakthrough curves studies and modeling. J. Hazard. Mater. 2011; 189(1-2):141-149.
- Mohammad HK. Biosorption of uranium from aqueous solutions by nonliving biomass of marine algae Cystoseria indica. Electron. J. Biotechnol. 2006; 9(2):100-106.
- Bhat SV, Melo JS, Chaugule BB. Biosorption characteristics of uranium(VI) from aqueous medium onto Catenella repens, a red alga. J. Hazard. Mater. 2008; 158(2-3):628-635.
- Zhang Z, Liu H, Song W. Accumulation of U(Ⅵ) on the Pantoea sp. TW18 isolated from radionuclide-contaminated soils. Journal of Environmental Radioactivity. 2018; 192(12):219-226.
- Wang Y, Nie X, Cheng W. A synergistic biosorption and biomineralization strategy for Kocuria sp. to immobilize U(VI) from aqueous solution. J. Mol. Liq. 2018; 275.
- Song W, Wang X, Sun Y. Bioaccumulation and transformation of U(VI) by sporangiospores of Mucor circinelloides. Chem. Eng. J. 2019; 362:81-88.
- Song W, Wang X, Chen Z. Enhanced immobilization of U(VI) on Mucor circinelloides in the presence of As(V): Batch and XAFS investigation. Environ. Pollut. 2018; 237(6):228-236.
- Wang J, Hu X, Liu Y. Biosorption of uranium (VI) by immobilized Aspergillus fumigatus beads. J. Environ. Radioact. 2010; 101(6):504-508.
- Sun Y, Zhang R, Ding C. Adsorption of U(VI) on sericite in the presence of Bacillus subtilis: A combined batch, EXAFS and modeling techniques. Geochim.Cosmochim.Acta. 2016; 180:51-65.
- Sivaswamy V, Boyanov MI, Peyton BM. Multiple mechanisms of uranium immobilization by <em>Cellulomonas</em> sp. strain ES6. Biotechnol. Bioeng. 2011; 108(2):264-276.
- Zou L, Chen Z, Zhang X. Phosphate promotes uranium (VI) adsorption in Staphylococcus aureus LZ‐01. Lett. Appl. Microbiol. 2014; 59.
- Zheng X, Wang X, Shen Y. Biosorption and biomineralization of uranium(VI) by Saccharomyces cerevisiae-Crystal formation of chernikovite. Chemosphere. 2017.
- Shen Y, Zheng X, Wang X. The biomineralization process of uranium(VI) by Saccharomyces cerevisiae-transformation from amorphous U(VI) to crystalline chernikovite. Appl.Microbiol.Biotechnol. 2018; 102(9):4217-4229.
- Sar P, Choudhary S. Interaction of uranium (VI) with bacteria: potential applications in bioremediation of U contaminated oxic environments. Rev.Environ.Sci.Biotechnol. 2015; 14(3):347-355.
- Sivaswamy V, Boyanov MI, Peyton BM. Multiple mechanisms of uranium immobilization by <em>Cellulomonas</em> sp. strain ES6. Biotechnology & Bioengineering. 2011; 108(2):264-276.
- Orellana R, Leavitt JJ, Comolli LR. U(VI) reduction by diverse outer surface c-type cytochromes of Geobacter sulfurreducens. Appl.Environ.Microbiol. 2013; 79(20):6369-6374.
- Li XL, Ding CC, Liao JL. Biosorption of uranium on Bacillus sp. dwc-2: preliminary investigation on mechanism. Journal of Environmental Radioactivity. 2014; 135:6-12.
- Liang X, Hillier S, Pendlowski H. Uranium phosphate biomineralization by fungi. Environ. Microbiol. 2015;17(6):2064-2075.
- Woolfolk CA, Whiteley HR. Reduction of inorganic compounds with molecular hydrogen by Micrococcus lactilyticus. I. Stoichiometry with compounds of arsenic, selenium, tellurium, transition, and other elements. J. Bacteriol. 1962; 84(4):647-658.
- Lovley DR, Phillips EJ. Microbial reduction of uranium. Nature. 1991.
- Lovley DR, Roden EE, Phillips EJ. Enzymatic iron and uranium reduction by sulfate-reducing bacteria. Mar.Geol. 1993; 113(1–2):41-53.
- Derek R, Lovley DR, Elizabeth J. Bioremediation of uranium contamination with enzymatic uranium reduction. Environ. Sci. Technol. 1992.
- Gorby YA, Lovley DR. Enzymic uranium precipitation. Environ.Sci. Technol. 1992; 26(1):205-207.
- Li X, Ding C, Liao J. Microbial reduction of uranium (VI) by Bacillus sp. dwc-2: A macroscopic and spectroscopic study. J. Environ. Sci. 2016; 53(3):9-15.
- Reguera G, Mccarthy K, Mehta T. Extracellular Electron Transfer Via Microbial Nanowires. Nature. 2005; 435(7045):1098-1101.
- Stirling CH, Andersen MB, Warthmann R. Isotope fractionation of 238U and 235U during biologically-mediated uranium reduction. Geochim. Cosmochim. Acta. 2015; 163:200-218.
- Zhang P, He Z, Van Nostrand JD. Dynamic Succession of Groundwater Sulfate-Reducing Communities during Prolonged Reduction of Uranium in a Contaminated Aquifer. J. Environ. Sci. Technol. 2017; 51(7):3609-3620.
- Wall JD, Krumholz LR. Uranium Reduction. Annual Review of Microbiology. 2006; 60(1):149-166.
- Bernier-Latmani R, Veeramani H, Vecchia ED. Non-uraninite Products of Microbial U(VI) Reduction. Environ. Sci. Technol. 2010; 44(24):9456-9462.
- Madden AS, Smith AC, Balkwill DL. Microbial uranium immobilization is independent of nitrate reduction. Environ. Microbiol. 2007; 9(9):2321-2330.
- Petrie L, North NN, Dollhopf SL. Enumeration and Characterization of Iron(III)-Reducing Microbial Communities from Acidic Subsurface Sediments Contaminated with Uranium(VI). Appl.Environ.Microbiol. 2003; 69(12):7467-7479.
- Dong W, Xie G, Miller TR. Sorption and bioreduction of hexavalent uranium at a military facility by the Chesapeake Bay. Environ. Pollut. 2006; 142(1):132-142.
- Tebo BM, Ya OA. Sulfate-reducing bacterium grows with Cr(VI), U(VI), Mn(IV), and Fe(III) as electron acceptors. FEMS Microbiol. Lett. 1998; 162(1):193-199.
- Junier P, Frutschi M, Wigginton NS, et al. Metal reduction by spores of Desulfotomaculum reducens. Environ. Microbiol. 2009; 11(12):3007-3017.
- Lloyd JR. Microbial reduction of metals and radionuclides. Fems Microbiol. Rev. (2003), 27(2-3):411-425.
- Mtimunye PJ, Chirwa EM. Characterization of the biochemical pathway of uranium (Ⅵ) reduction in facultative anaerobic bacteria. Chemosphere. 2014; 113:22-29.
- Shelobolina ES, Sullivan SA, O'Neill KR. Isolation, characterization, and U(VI) reducing potential of a facultatively anaerobic, acid-resistant bacterium from low pH, nitrate, and U(VI) contaminated subsurface sediment and description of Salmonella subterranea sp. Appl. Environ. Microbiol. 2004; 70(5):2959-2965.
- Ganesh R, Robinson KG, Reed GD. Reduction of hexavalent uranium from organic complexes by sulfate- and iron-reducing bacteria. Appl. Environ. Microbiol. 1997; 63(11):4385-4391.
- Ling S, Szymanowski J, Fein JB. The effects of uranium speciation on the rate of U(VI) reduction by Shewanella oneidensis MR-1. Geochim. Cosmochim. Acta. 2011; 75(12):3558-3567.
- Jiang S, Kim MG, Kim SJ. Bacterial formation of extracellular U(VI) nanowires. Chem. Commun. 2011; 47(28):8076-8078.
- Renshaw JC, Butchins LJ, Livens FR. Bioreduction of uranium: Environmental implications of a pentavalent intermediate. Environ. Sci. Technol. 2005; 39(15):5657-5660.
- Senko JM, Kelly SD, Dohnalkova AC. The effect of U(VI) bioreduction kinetics on subsequent reoxidation of biogenic U(IV). Geochim. Cosmochim. Acta. 2007; 71(19):0-4654.
- Suzuki Y, Kelly SD, Kemner KM. Radionuclide contamination: Nanometre-size products of uranium bioreduction. Nature. 2002; 419(6903):134-134.
- Bargar JR, Bernier-Latmani R, Giammar DE. Biogenic Uraninite Nanoparticles and Their Importance for Uranium Remediation. Elements. 2008; 4(6):407-412.
- Marshall MJ, Beliaev AS, Dohnalkova AC. c-Type Cytochrome-Dependent Formation of U(IV) Nanoparticles by Shewanella oneidensis. PLoS Biol. 2006; 4(8):1324-1333.
- Shi L, Belchik SM, Wang Z. Identification and Characterization of UndAHRCR-6, an Outer Membrane Endecaheme c-Type Cytochrome of Shewanella sp. Strain HRCR-6. Appl. Environ. Microbiol. 2011; 77(15):5521-5523.
- Cao B, Ahmed B, Kennedy DW, et al. Contribution of Extracellular Polymeric Substances from Shewanella sp. HRCR-1 Biofilms to U(VI) Immobilization. Environ. Sci. Technol. (2011), 45(13):5483-5490.
- Sani RK, Peyton BM, Dohnalkova A. Comparison of uranium(Ⅵ) removal by Shewanella oneidensis MR-1 in flow and batch reactors. Water Res. 2008; 42(12):2993-3002.
- Onuki T. Flavin mononucleotide mediated reduction of U(VI). Geochim. Cosmochim. Acta. 2009; 73(13):A1298-A1298.
- Finneran KT, Housewright ME, Lovley DR. Multip, le influences of nitrate on uranium solubility durin~1 bioremediation of uranium-contaminated subsurface sediments. Environ Microbiol. 2002; 4(9):510-516.
- Cologgi DL, Lampa-Pastirk S, Speers AM. Extracellular reduction of uranium via Geobacter conductive pili as a protective cellular mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2011; 108(37):15248-15252.
- Reguera G. Electron transfer at the cell-uranium interface in Geobacter spp. Biochem.Soc.Trans. 2012; 40(6):1227-1232.
- Sanford RA, Wu Q, Sung Y. Hexavalent uranium supports the growth of Anaeromyxobacter dehalogenans and Geobacter spp. with lower-than-predicted biomass yields. Environ. Microbiol. 2007; 9(11):2885-2893.
- Liu N, Li F, Yang Y. Biosorption and bioaccumulation behavior of uranium on Bacillus sp dwc-2: Investigation by Box-Behenken design method. J. Mol. Liq. 2016; 221:156-165.
- Guenay A, Arslankaya E, Tosun I. Lead removal from aqueous solution by natural and pretreated clinoptilolite: Adsorption equilibrium and kinetics. J. Hazard. Mater. 2007; 146(1-2):362-371.
- Luo W, Wu WM, Yan T. Influence of bicarbonate, sulfate, and electron donors on biological reduction of uranium and microbial community composition. Appl. Microbiol. Biotechnol. 2007; 77(3):713-721.
- Ortiz-Bernad I, Anderson RT, Vrionis HA. Resistance of solid-phase U(VI) to microbial reduction during in situ bioremediation of uranium-contaminated groundwater. Appl. Environ. Microbiol. 2004; 70(12):7558-7560.
- Wu QZ, Sanford RA, Löffler FE. Uranium(VI) Reduction by Anaeromyxobacter dehalogenans Strain 2CP-C. Appl. Environ. Microbiol. 2006; 72(5):3608-3614.
- Phillips EJ, Landa ER, Lovley DR. Remediation of uranium contaminated soils with bicarbonate extraction and microbial U(VI) reduction. J. Ind. Microbiol. 1995; 14(3-4):203-207.
- Istok JD, Senko JM, Krumholz LR. In Situ Bioreduction of Technetium and Uranium in a Nitrate-Contaminated Aquifer. Environ. Sci. Technol. 2004; 38(2):468-475.
- Williams KH, Long PE, Davis JA. Acetate Availability and its Influence on Sustainable Bioremediation of Uranium-Contaminated Groundwater. Geomicrobiol. J. 2011; 28(5-6):519-539.
- Barlett M, Moon HS, Peacock AA, et al. Uranium reduction and microbial community development in response to stimulation with different electron donors. Biodegradation. 2012; 23(4):535-546.
- Watson DB, Wu WM, Mehlhorn T. In Situ Bioremediation of Uranium with Emulsified Vegetable Oil as the Electron Donor. J. Environ. Sci. Technol. 2013; 47(12).
- Li B, Wu W, Watson DB. Bacterial community shift and coexisting/co-excluding patterns revealed by network analysis in a bio-reduced uranium-contaminated site after reoxidation. Appl. Environ. Microbiol. 2018; 84(9).
- Merroun ML, Nedelkova M, Ojeda JJ. Bio-precipitation of uranium by two bacterial isolates recovered from extreme environments as estimated by potentiometric titration, TEM, and X-ray absorption spectroscopic analyses. J. Hazard. Mater. 2011; 197:1-10.
- Kapoor A, Viraraghavan T. Heavy metal biosorption sites in Aspergillus niger. Bioresour. Technol. 1997; 61(3):221-227.
- Islam E, Sar P. Diversity, metal resistance and uranium sequestration abilities of bacteria from uranium ore deposit in deep earth stratum. Ecotoxicology & Environmental Safety. 2016; 127:12-21.
- Zheng XY, Lu X, Wang XY. Biosorption and biomineralization of uranium(VI) by Saccharomyces cerevisiae-Crystal formation of chernikovite.Chemosphere Environmental Toxicology & Risk Assessment. 2017.
- Martinez RJ, Beazley MJ, Taillefert M. Aerobic uranium (VI) bio-precipitation by metal-resistant bacteria isolated from radionuclide- and metal-contaminated subsurface soils. Environ. Microbiol. 2007; 9(12):3122-3133.
- Salome KR, Beazley MJ, Webb SM. Biomineralization of U(VI) phosphate promoted by microbially-mediated phytate hydrolysis in contaminated soils. Geochim. Cosmochim. Acta. 2017; 197:27-42.
- Bader M, Mueller K, Foerstendorf H. Multistage bioassociation of uranium onto an extremely halophilic archaeon revealed by a unique combination of spectroscopic and microscopic techniques. J. Hazard. Mater. 2017; 327:225-232.
- Gerber U, Zirnstein I, Krawczyk-Baersch E. Combined use of flow cytometry and microscopy to study the interactions between. the gram-negative beta proteobacterium Acidovorax facilis and uranium(VI). J. Hazard. Mater. 2016; 317:127-134.
- Pan X, Chen Z, Chen F, et al. The mechanism of uranium transformation from U(VI) into nano-uramphite by two indigenous Bacillus thuringiensis strains. J. Hazard. Mater. 2015; 297:313-319.
- Nilesh K, Smita Z, Celin A. Responses exhibited by various microbial groups relevant to uranium exposure.Biotechnol. Adv.: An International Review Journal. 2018; 36(7).
- Huang W, Cheng W, Nie X. AlharbiMicroscopic and Spectroscopic Insights into Uranium Phosphate Mineral Precipitated by Bacillus Mucilaginosus. ACS Earth Space Chem. 2017; 1:483-492.
- Mukherjee A, Wheaton GH, Blum PH. Uranium extremophile is an adaptive, rather than intrinsic, feature for extremely thermoacidophilic Metallosphaera species. Proceedings of the National Academy of Sciences of the United States of America. 2012; 109(41): 6.
- Holmes DE, Nevin KP, O'Neil RA, et al. Potential for Quantifying Expression of the Geobacteraceae in the Subsurface and on Current-Harvesting Electrodes. Appl Environ Microbiol. 2005; 71(11):6870-6877.
- Wilkins MJ, Callister SJ, Miletto M. Development of a biomarker for Geobacter activity and strain composition; Proteogenomic analysis of the citrate synthase protein during bioremediation of U(VI). Microb. Biotechnol. 2010; 4(1):55-63.
- Bruinenberg PM, Van Dijken JP, Scheffers WA. An Enzymic Analysis of NADPH Production and Consumption in Candida utilis. J. Gen. Microbiol. 1983; 129(4):965-971.
- Song W, Liang J, Tao W. Accumulation of Co(II) and Eu(III) by the mycelia of Aspergillus niger isolated from radionuclide-contaminated soils. Chem. Eng. J. 2016; 304:186-193.
- Orellana R, Hixson KK, Murphy S. Proteome of Geobacter sulfurreducens in the presence of U(VI). Microbiol. 2014; 160:2607-2617.
- Kim EH, Nies DH, McEvoy MM, Rensing C. Switch or funnel: how RND-type transport systems control periplasmic metal homeostasis. J Bacteriol. 2011 May;193(10):2381-7. doi: 10.1128/JB.01323-10. Epub 2011 Mar 11. PMID: 21398536; PMCID: PMC3133179.
- Orellana R, Hixson KK, Murphy S, Mester T, Sharma ML, Lipton MS, Lovley DR. Proteome of Geobacter sulfurreducens in the presence of U(VI). Microbiology (Reading). 2014 Dec;160(Pt 12):2607-2617. doi: 10.1099/mic.0.081398-0. Epub 2014 Oct 1. PMID: 25273002.
- Nicolas G, Béatrice AB, Philippe O. Proteogenomic insights into uranium tolerance of a Chernobyl's Microbacterium bacterial isolate. J. Proteomics. 2017; 177:148-157.
- Andrews SC, Robinson AK, Francisco RQ. Bacterial iron homeostasis. Fems Microbiology Reviews. 2003; (2-3):2-3.
- Krushkal J, Sontineni S, Leang C, Qu Y, Adkins RM, Lovley DR. Genome diversity of the TetR family of transcriptional regulators in a metal-reducing bacterial family Geobacteraceae and other microbial species. OMICS. 2011 Jul-Aug;15(7-8):495-506. doi: 10.1089/omi.2010.0117. Epub 2011 Jun 23. PMID: 21699403.
- Summers AO. Damage control: regulating defenses against toxic metals and metalloids. Curr. Opin. Microbiol. 2009; 12(2):138-144.
- Tottey S, Harvie DR, Robinson NJ. Understanding How Cells Allocate Metals Using Metal Sensors and Metallochaperones. Acc. Chem. Res. 2005; 37(3).
- Ueki T, Lovley DR. Heat-Shock Sigma Factor RpoH from Geobacter Sulfurreducens. Microbiol. 2007; 153:838-846.
- Hu P, Yohey H. Whole-Genome Transcriptional Analysis of Heavy Metal Stresses in Caulobacter crescentus. J. Bacteriol. 2005; 187(24):8437-8449.
- Lloyd JR, Lovley DR. Microbial detoxification of metals and radionuclides. Curr.Opin.Biotechnol. 2001; 12(3):248-253.
- Seufferheld MJ, Alvarez HM, Farias ME. Role of Polyphosphates in Microbial Adaptation to Extreme Environments. Appl. Environ. Microbiol. 2008; 74(19):5867-5874.
- Keasling JD, Hupf GA. Genetic manipulation of polyphosphate metabolism affects cadmium tolerance in Escherichia coli. Appl. Environ. Microbiol. 1996; 62(2):743-746.
- Keyhani S, Lopez JL, Clark DS. Intracellular polyphosphate content and cadmium tolerance in Anacystis nidulans R2. Microbios. 1996; 88(355):105-114.
- Yung MC, Ma J, Salemi MR. Shotgun Proteomic Analysis Unveils Survival and Detoxification Strategies by Caulobacter crescentus during Exposure to Uranium, Chromium, and Cadmium. J.Proteome Res. 2014; 13(4):1833-1847.
- Yung MC. Biomineralization of uranium by PhoY phosphatase activity aids cell survival in Caulobacter crescentus. Appl. Environ. Microbiol. 2014; 80(16):4795-804.
- Dekker L, Arsène-Ploetze F, Santini JM. Comparative proteomics of AcidithioBacillus ferrooxidans grown in the presence and absence of uranium. Res. Microbiol. 2016; 167(3):234-239.
- Khemiri A, Carrière M, Bremond N. Escherichia coli Response to Uranyl Exposure at Low pH and Associated Protein Regulations. Plos One. 2014; 9(2).
- Matsuda E, Nakajima A. Effect of catechins and tannins on depleted uranium-induced DNA strand breaks. J. Radioanal. Nucl. Chem. 2012; 293(2):711-714.
- Ghosh M, Rosen B. Microbial Resistance Mechanisms for Heavy Metals and Metalloids. Nd Ecker. 2002.
- Yazzie M, Gamble SL, Civitello ER. Uranyl Acetate Causes DNA Single-Strand Breaks In Vitro in the Presence of Ascorbate (Vitamin C). Chem. Res. Toxicol. 2003; 16(4):524-530.
- Zobel CR, Beer M. Electron Stains: I. Chemical Studies on the Interaction of DNA with Uranyl Salts. The Journal of Biophysical and Biochemical Cytology. 1961; 10(3):335-346.
- Horn DV, Huang H. Uranium(VI) bio-coordination chemistry from biochemical, solution and protein structural data. Coord. Chem. Rev. 2006; 250(7-8):765-775.
- Lovley DR, Ueki T, Zhang T. Geobacter: The Microbe Electric's Physiology, Ecology, and Practical Applications. Adv. Microb.Physiol. 2011; 59:1-100.
- Teena M, Susan EC, Richard G. A putative multicopper protein secreted by an atypical type II secretion system involved in the reduction of insoluble electron acceptors in Geobacter sulfurreducens. Microbiol. 2006; 152(8):2257-2264.
- Hartsock WJ, Cohen JD, Segal DJ. Uranyl Acetate is a Direct Inhibitor of DNA-Binding Proteins. Chem. Res. Toxicol. 2007; 20(5):784-789.
- Mouser PJ, Holmes DE, Perpetua LA. Quantifying expression of Geobacter spp. oxidative stress genes in pure culture and during in situ uranium bioremediation. Isme Journal. 2009; 3(4):454-465.
- Shelobolina ES, Vrionis HA, Findlay RH. Geobacter uraniireducens sp. nov., isolated from subsurface sediment undergoing uranium bioremediation. International Journal of Systematic & Evolutionary Microbiology. (2008); 58(5):1075-1078.
- Ping Hu, Brodie E, Suzuki Y. Whole-Genome Transcriptional Analysis of Heavy Metal Stresses in Caulobacter crescentus. Journal of Bacteriology. 2005.

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.
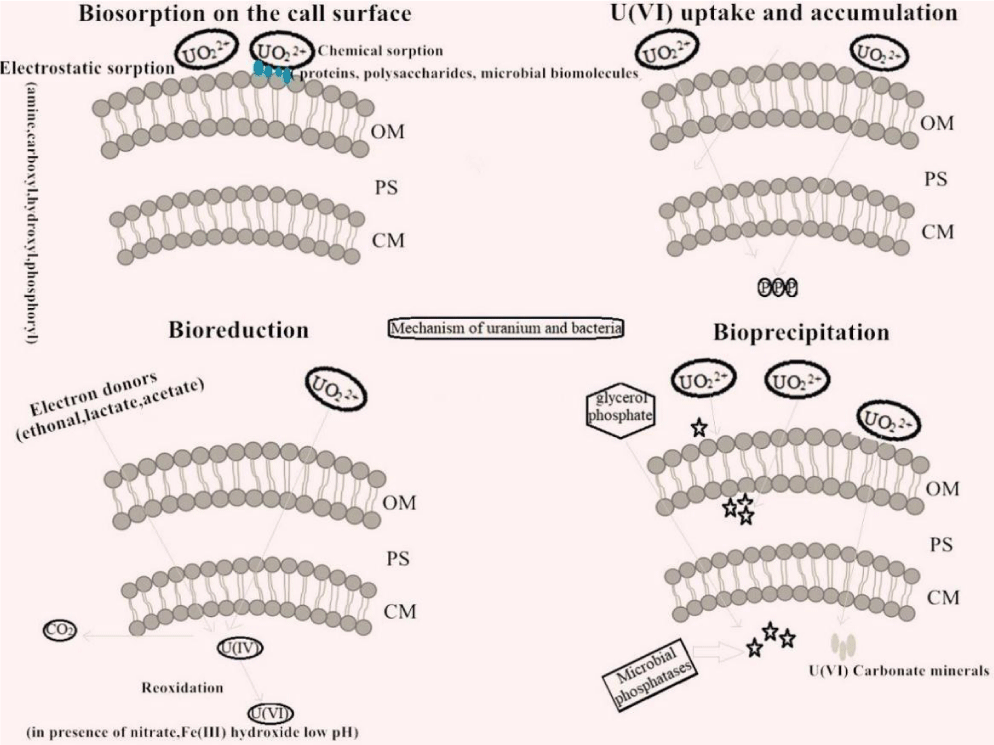

 Save to Mendeley
Save to Mendeley
